Chi-en Amy Tai
Cancer-Net PCa-MultiSeg: Multimodal Enhancement of Prostate Cancer Lesion Segmentation Using Synthetic Correlated Diffusion Imaging
Nov 11, 2025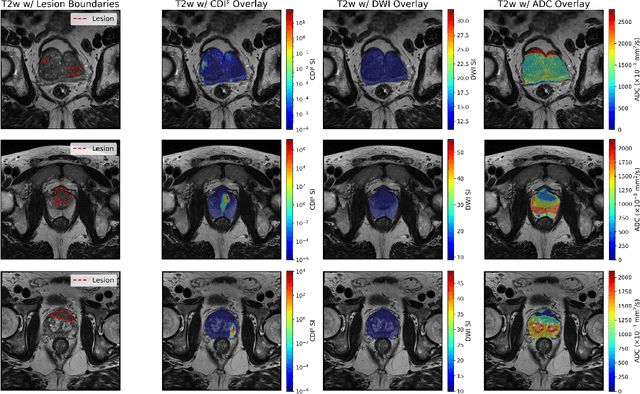
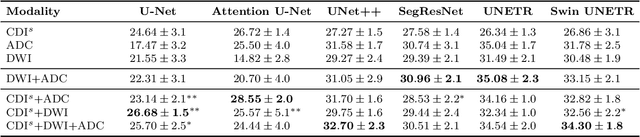
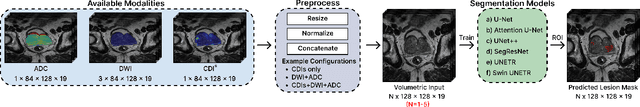
Abstract:Current deep learning approaches for prostate cancer lesion segmentation achieve limited performance, with Dice scores of 0.32 or lower in large patient cohorts. To address this limitation, we investigate synthetic correlated diffusion imaging (CDI$^s$) as an enhancement to standard diffusion-based protocols. We conduct a comprehensive evaluation across six state-of-the-art segmentation architectures using 200 patients with co-registered CDI$^s$, diffusion-weighted imaging (DWI) and apparent diffusion coefficient (ADC) sequences. We demonstrate that CDI$^s$ integration reliably enhances or preserves segmentation performance in 94% of evaluated configurations, with individual architectures achieving up to 72.5% statistically significant relative improvement over baseline modalities. CDI$^s$ + DWI emerges as the safest enhancement pathway, achieving significant improvements in half of evaluated architectures with zero instances of degradation. Since CDI$^s$ derives from existing DWI acquisitions without requiring additional scan time or architectural modifications, it enables immediate deployment in clinical workflows. Our results establish validated integration pathways for CDI$^s$ as a practical drop-in enhancement for PCa lesion segmentation tasks across diverse deep learning architectures.
Clinical trial cohort selection using Large Language Models on n2c2 Challenges
Jan 19, 2025



Abstract:Clinical trials are a critical process in the medical field for introducing new treatments and innovations. However, cohort selection for clinical trials is a time-consuming process that often requires manual review of patient text records for specific keywords. Though there have been studies on standardizing the information across the various platforms, Natural Language Processing (NLP) tools remain crucial for spotting eligibility criteria in textual reports. Recently, pre-trained large language models (LLMs) have gained popularity for various NLP tasks due to their ability to acquire a nuanced understanding of text. In this paper, we study the performance of large language models on clinical trial cohort selection and leverage the n2c2 challenges to benchmark their performance. Our results are promising with regard to the incorporation of LLMs for simple cohort selection tasks, but also highlight the difficulties encountered by these models as soon as fine-grained knowledge and reasoning are required.
Cancer-Net PCa-Seg: Benchmarking Deep Learning Models for Prostate Cancer Segmentation Using Synthetic Correlated Diffusion Imaging
Jan 15, 2025


Abstract:Prostate cancer (PCa) is the most prevalent cancer among men in the United States, accounting for nearly 300,000 cases, 29% of all diagnoses and 35,000 total deaths in 2024. Traditional screening methods such as prostate-specific antigen (PSA) testing and magnetic resonance imaging (MRI) have been pivotal in diagnosis, but have faced limitations in specificity and generalizability. In this paper, we explore the potential of enhancing PCa lesion segmentation using a novel MRI modality called synthetic correlated diffusion imaging (CDI$^s$). We employ several state-of-the-art deep learning models, including U-Net, SegResNet, Swin UNETR, Attention U-Net, and LightM-UNet, to segment PCa lesions from a 200 CDI$^s$ patient cohort. We find that SegResNet achieved superior segmentation performance with a Dice-Sorensen coefficient (DSC) of $76.68 \pm 0.8$. Notably, the Attention U-Net, while slightly less accurate (DSC $74.82 \pm 2.0$), offered a favorable balance between accuracy and computational efficiency. Our findings demonstrate the potential of deep learning models in improving PCa lesion segmentation using CDI$^s$ to enhance PCa management and clinical support.
Cancer-Net SCa-Synth: An Open Access Synthetically Generated 2D Skin Lesion Dataset for Skin Cancer Classification
Nov 08, 2024



Abstract:In the United States, skin cancer ranks as the most commonly diagnosed cancer, presenting a significant public health issue due to its high rates of occurrence and the risk of serious complications if not caught early. Recent advancements in dataset curation and deep learning have shown promise in quick and accurate detection of skin cancer. However, current open-source datasets have significant class imbalances which impedes the effectiveness of these deep learning models. In healthcare, generative artificial intelligence (AI) models have been employed to create synthetic data, addressing data imbalance in datasets by augmenting underrepresented classes and enhancing the overall quality and performance of machine learning models. In this paper, we build on top of previous work by leveraging new advancements in generative AI, notably Stable Diffusion and DreamBooth. We introduce Cancer-Net SCa-Synth, an open access synthetically generated 2D skin lesion dataset for skin cancer classification. Further analysis on the data effectiveness by comparing the ISIC 2020 test set performance for training with and without these synthetic images for a simple model highlights the benefits of leveraging synthetic data to improve performance. Cancer-Net SCa-Synth is publicly available at https://github.com/catai9/Cancer-Net-SCa-Synth as part of a global open-source initiative for accelerating machine learning for cancer care.
Enhancing Trust in Clinically Significant Prostate Cancer Prediction with Multiple Magnetic Resonance Imaging Modalities
Nov 07, 2024



Abstract:In the United States, prostate cancer is the second leading cause of deaths in males with a predicted 35,250 deaths in 2024. However, most diagnoses are non-lethal and deemed clinically insignificant which means that the patient will likely not be impacted by the cancer over their lifetime. As a result, numerous research studies have explored the accuracy of predicting clinical significance of prostate cancer based on magnetic resonance imaging (MRI) modalities and deep neural networks. Despite their high performance, these models are not trusted by most clinical scientists as they are trained solely on a single modality whereas clinical scientists often use multiple magnetic resonance imaging modalities during their diagnosis. In this paper, we investigate combining multiple MRI modalities to train a deep learning model to enhance trust in the models for clinically significant prostate cancer prediction. The promising performance and proposed training pipeline showcase the benefits of incorporating multiple MRI modalities for enhanced trust and accuracy.
Using Multiparametric MRI with Optimized Synthetic Correlated Diffusion Imaging to Enhance Breast Cancer Pathologic Complete Response Prediction
May 13, 2024

Abstract:In 2020, 685,000 deaths across the world were attributed to breast cancer, underscoring the critical need for innovative and effective breast cancer treatment. Neoadjuvant chemotherapy has recently gained popularity as a promising treatment strategy for breast cancer, attributed to its efficacy in shrinking large tumors and leading to pathologic complete response. However, the current process to recommend neoadjuvant chemotherapy relies on the subjective evaluation of medical experts which contain inherent biases and significant uncertainty. A recent study, utilizing volumetric deep radiomic features extracted from synthetic correlated diffusion imaging (CDI$^s$), demonstrated significant potential in noninvasive breast cancer pathologic complete response prediction. Inspired by the positive outcomes of optimizing CDI$^s$ for prostate cancer delineation, this research investigates the application of optimized CDI$^s$ to enhance breast cancer pathologic complete response prediction. Using multiparametric MRI that fuses optimized CDI$^s$ with diffusion-weighted imaging (DWI), we obtain a leave-one-out cross-validation accuracy of 93.28%, over 5.5% higher than that previously reported.
Improving Breast Cancer Grade Prediction with Multiparametric MRI Created Using Optimized Synthetic Correlated Diffusion Imaging
May 13, 2024



Abstract:Breast cancer was diagnosed for over 7.8 million women between 2015 to 2020. Grading plays a vital role in breast cancer treatment planning. However, the current tumor grading method involves extracting tissue from patients, leading to stress, discomfort, and high medical costs. A recent paper leveraging volumetric deep radiomic features from synthetic correlated diffusion imaging (CDI$^s$) for breast cancer grade prediction showed immense promise for noninvasive methods for grading. Motivated by the impact of CDI$^s$ optimization for prostate cancer delineation, this paper examines using optimized CDI$^s$ to improve breast cancer grade prediction. We fuse the optimized CDI$^s$ signal with diffusion-weighted imaging (DWI) to create a multiparametric MRI for each patient. Using a larger patient cohort and training across all the layers of a pretrained MONAI model, we achieve a leave-one-out cross-validation accuracy of 95.79%, over 8% higher compared to that previously reported.
Optimizing Synthetic Correlated Diffusion Imaging for Breast Cancer Tumour Delineation
May 13, 2024


Abstract:Breast cancer is a significant cause of death from cancer in women globally, highlighting the need for improved diagnostic imaging to enhance patient outcomes. Accurate tumour identification is essential for diagnosis, treatment, and monitoring, emphasizing the importance of advanced imaging technologies that provide detailed views of tumour characteristics and disease. Synthetic correlated diffusion imaging (CDI$^s$) is a recent method that has shown promise for prostate cancer delineation compared to current MRI images. In this paper, we explore tuning the coefficients in the computation of CDI$^s$ for breast cancer tumour delineation by maximizing the area under the receiver operating characteristic curve (AUC) using a Nelder-Mead simplex optimization strategy. We show that the best AUC is achieved by the CDI$^s$ - Optimized modality, outperforming the best gold-standard modality by 0.0044. Notably, the optimized CDI$^s$ modality also achieves AUC values over 0.02 higher than the Unoptimized CDI$^s$ value, demonstrating the importance of optimizing the CDI$^s$ exponents for the specific cancer application.
NutritionVerse-Direct: Exploring Deep Neural Networks for Multitask Nutrition Prediction from Food Images
May 13, 2024



Abstract:Many aging individuals encounter challenges in effectively tracking their dietary intake, exacerbating their susceptibility to nutrition-related health complications. Self-reporting methods are often inaccurate and suffer from substantial bias; however, leveraging intelligent prediction methods can automate and enhance precision in this process. Recent work has explored using computer vision prediction systems to predict nutritional information from food images. Still, these methods are often tailored to specific situations, require other inputs in addition to a food image, or do not provide comprehensive nutritional information. This paper aims to enhance the efficacy of dietary intake estimation by leveraging various neural network architectures to directly predict a meal's nutritional content from its image. Through comprehensive experimentation and evaluation, we present NutritionVerse-Direct, a model utilizing a vision transformer base architecture with three fully connected layers that lead to five regression heads predicting calories (kcal), mass (g), protein (g), fat (g), and carbohydrates (g) present in a meal. NutritionVerse-Direct yields a combined mean average error score on the NutritionVerse-Real dataset of 412.6, an improvement of 25.5% over the Inception-ResNet model, demonstrating its potential for improving dietary intake estimation accuracy.
Enhancing Clinically Significant Prostate Cancer Prediction in T2-weighted Images through Transfer Learning from Breast Cancer
May 13, 2024


Abstract:In 2020, prostate cancer saw a staggering 1.4 million new cases, resulting in over 375,000 deaths. The accurate identification of clinically significant prostate cancer is crucial for delivering effective treatment to patients. Consequently, there has been a surge in research exploring the application of deep neural networks to predict clinical significance based on magnetic resonance images. However, these networks demand extensive datasets to attain optimal performance. Recently, transfer learning emerged as a technique that leverages acquired features from a domain with richer data to enhance the performance of a domain with limited data. In this paper, we investigate the improvement of clinically significant prostate cancer prediction in T2-weighted images through transfer learning from breast cancer. The results demonstrate a remarkable improvement of over 30% in leave-one-out cross-validation accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge