Xavier Tannier
LIMICS
SynCABEL: Synthetic Contextualized Augmentation for Biomedical Entity Linking
Jan 27, 2026Abstract:We present SynCABEL (Synthetic Contextualized Augmentation for Biomedical Entity Linking), a framework that addresses a central bottleneck in supervised biomedical entity linking (BEL): the scarcity of expert-annotated training data. SynCABEL leverages large language models to generate context-rich synthetic training examples for all candidate concepts in a target knowledge base, providing broad supervision without manual annotation. We demonstrate that SynCABEL, when combined with decoder-only models and guided inference establish new state-of-the-art results across three widely used multilingual benchmarks: MedMentions for English, QUAERO for French, and SPACCC for Spanish. Evaluating data efficiency, we show that SynCABEL reaches the performance of full human supervision using up to 60% less annotated data, substantially reducing reliance on labor-intensive and costly expert labeling. Finally, acknowledging that standard evaluation based on exact code matching often underestimates clinically valid predictions due to ontology redundancy, we introduce an LLM-as-a-judge protocol. This analysis reveals that SynCABEL significantly improves the rate of clinically valid predictions. Our synthetic datasets, models, and code are released to support reproducibility and future research.
Development of the user-friendly decision aid Rule-based Evaluation and Support Tool (REST) for optimizing the resources of an information extraction task
Jun 16, 2025Abstract:Rules could be an information extraction (IE) default option, compared to ML and LLMs in terms of sustainability, transferability, interpretability, and development burden. We suggest a sustainable and combined use of rules and ML as an IE method. Our approach starts with an exhaustive expert manual highlighting in a single working session of a representative subset of the data corpus. We developed and validated the feasibility and the performance metrics of the REST decision tool to help the annotator choose between rules as a by default option and ML for each entity of an IE task. REST makes the annotator visualize the characteristics of each entity formalization in the free texts and the expected rule development feasibility and IE performance metrics. ML is considered as a backup IE option and manual annotation for training is therefore minimized. The external validity of REST on a 12-entity use case showed good reproducibility.
Clinical trial cohort selection using Large Language Models on n2c2 Challenges
Jan 19, 2025



Abstract:Clinical trials are a critical process in the medical field for introducing new treatments and innovations. However, cohort selection for clinical trials is a time-consuming process that often requires manual review of patient text records for specific keywords. Though there have been studies on standardizing the information across the various platforms, Natural Language Processing (NLP) tools remain crucial for spotting eligibility criteria in textual reports. Recently, pre-trained large language models (LLMs) have gained popularity for various NLP tasks due to their ability to acquire a nuanced understanding of text. In this paper, we study the performance of large language models on clinical trial cohort selection and leverage the n2c2 challenges to benchmark their performance. Our results are promising with regard to the incorporation of LLMs for simple cohort selection tasks, but also highlight the difficulties encountered by these models as soon as fine-grained knowledge and reasoning are required.
Prompt engineering paradigms for medical applications: scoping review and recommendations for better practices
May 02, 2024



Abstract:Prompt engineering is crucial for harnessing the potential of large language models (LLMs), especially in the medical domain where specialized terminology and phrasing is used. However, the efficacy of prompt engineering in the medical domain remains to be explored. In this work, 114 recent studies (2022-2024) applying prompt engineering in medicine, covering prompt learning (PL), prompt tuning (PT), and prompt design (PD) are reviewed. PD is the most prevalent (78 articles). In 12 papers, PD, PL, and PT terms were used interchangeably. ChatGPT is the most commonly used LLM, with seven papers using it for processing sensitive clinical data. Chain-of-Thought emerges as the most common prompt engineering technique. While PL and PT articles typically provide a baseline for evaluating prompt-based approaches, 64% of PD studies lack non-prompt-related baselines. We provide tables and figures summarizing existing work, and reporting recommendations to guide future research contributions.
A Benchmark Evaluation of Clinical Named Entity Recognition in French
Mar 28, 2024



Abstract:Background: Transformer-based language models have shown strong performance on many Natural LanguageProcessing (NLP) tasks. Masked Language Models (MLMs) attract sustained interest because they can be adaptedto different languages and sub-domains through training or fine-tuning on specific corpora while remaining lighterthan modern Large Language Models (LLMs). Recently, several MLMs have been released for the biomedicaldomain in French, and experiments suggest that they outperform standard French counterparts. However, nosystematic evaluation comparing all models on the same corpora is available. Objective: This paper presentsan evaluation of masked language models for biomedical French on the task of clinical named entity recognition.Material and methods: We evaluate biomedical models CamemBERT-bio and DrBERT and compare them tostandard French models CamemBERT, FlauBERT and FrALBERT as well as multilingual mBERT using three publicallyavailable corpora for clinical named entity recognition in French. The evaluation set-up relies on gold-standardcorpora as released by the corpus developers. Results: Results suggest that CamemBERT-bio outperformsDrBERT consistently while FlauBERT offers competitive performance and FrAlBERT achieves the lowest carbonfootprint. Conclusion: This is the first benchmark evaluation of biomedical masked language models for Frenchclinical entity recognition that compares model performance consistently on nested entity recognition using metricscovering performance and environmental impact.
Few shot clinical entity recognition in three languages: Masked language models outperform LLM prompting
Feb 20, 2024



Abstract:Large Language Models are becoming the go-to solution for many natural language processing tasks, including in specialized domains where their few-shot capacities are expected to yield high performance in low-resource settings. Herein, we aim to assess the performance of Large Language Models for few shot clinical entity recognition in multiple languages. We evaluate named entity recognition in English, French and Spanish using 8 in-domain (clinical) and 6 out-domain gold standard corpora. We assess the performance of 10 auto-regressive language models using prompting and 16 masked language models used for text encoding in a biLSTM-CRF supervised tagger. We create a few-shot set-up by limiting the amount of annotated data available to 100 sentences. Our experiments show that although larger prompt-based models tend to achieve competitive F-measure for named entity recognition outside the clinical domain, this level of performance does not carry over to the clinical domain where lighter supervised taggers relying on masked language models perform better, even with the performance drop incurred from the few-shot set-up. In all experiments, the CO2 impact of masked language models is inferior to that of auto-regressive models. Results are consistent over the three languages and suggest that few-shot learning using Large language models is not production ready for named entity recognition in the clinical domain. Instead, models could be used for speeding-up the production of gold standard annotated data.
Impact of translation on biomedical information extraction from real-life clinical notes
Jun 03, 2023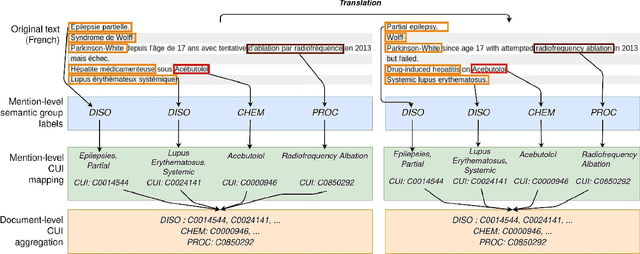
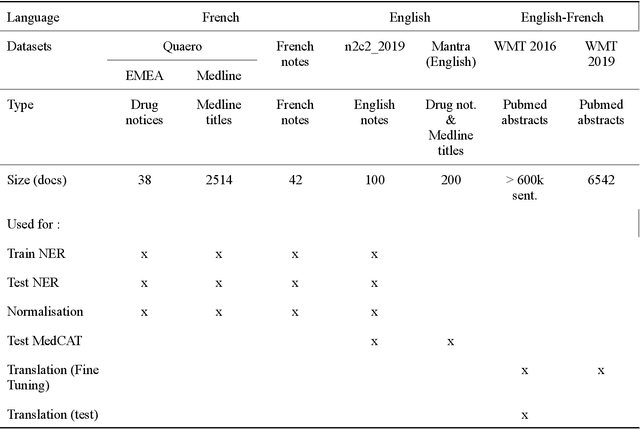
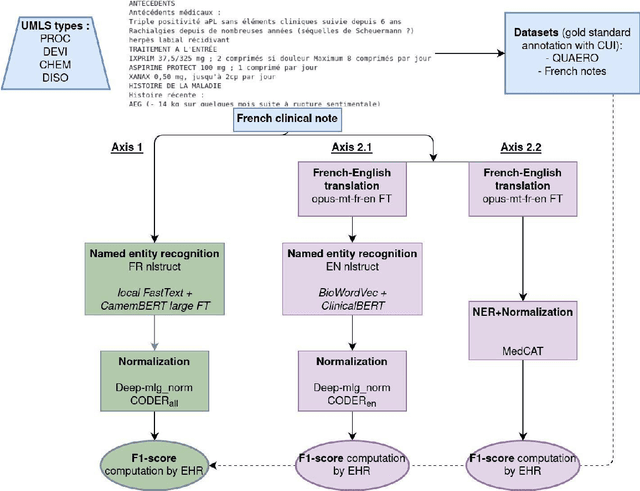
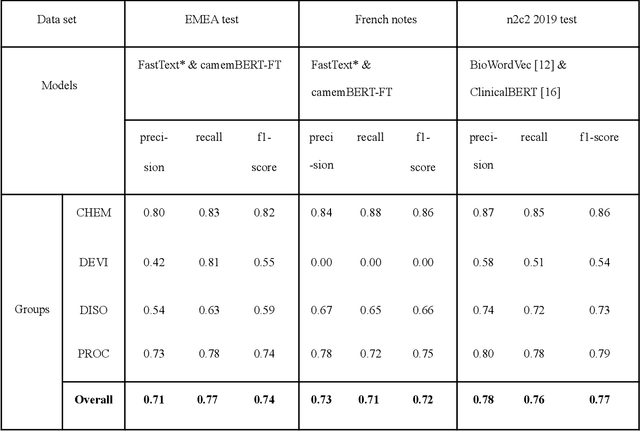
Abstract:The objective of our study is to determine whether using English tools to extract and normalize French medical concepts on translations provides comparable performance to French models trained on a set of annotated French clinical notes. We compare two methods: a method involving French language models and a method involving English language models. For the native French method, the Named Entity Recognition (NER) and normalization steps are performed separately. For the translated English method, after the first translation step, we compare a two-step method and a terminology-oriented method that performs extraction and normalization at the same time. We used French, English and bilingual annotated datasets to evaluate all steps (NER, normalization and translation) of our algorithms. Concerning the results, the native French method performs better than the translated English one with a global f1 score of 0.51 [0.47;0.55] against 0.39 [0.34;0.44] and 0.38 [0.36;0.40] for the two English methods tested. In conclusion, despite the recent improvement of the translation models, there is a significant performance difference between the two approaches in favor of the native French method which is more efficient on French medical texts, even with few annotated documents.
Detecting automatically the layout of clinical documents to enhance the performances of downstream natural language processing
May 23, 2023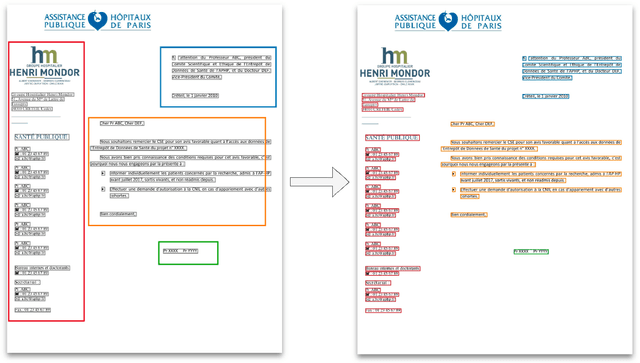


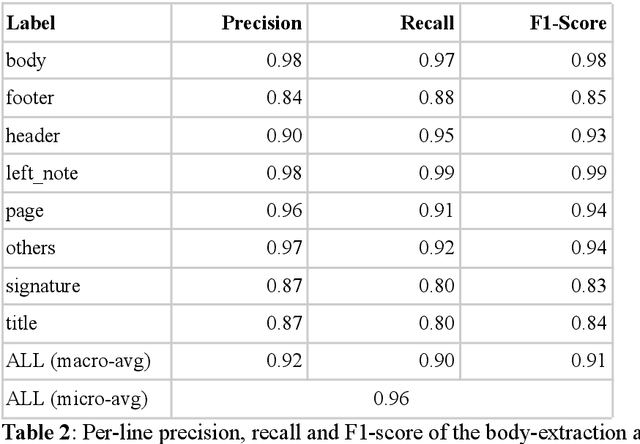
Abstract:Objective:Develop and validate an algorithm for analyzing the layout of PDF clinical documents to improve the performance of downstream natural language processing tasks. Materials and Methods: We designed an algorithm to process clinical PDF documents and extract only clinically relevant text. The algorithm consists of several steps: initial text extraction using a PDF parser, followed by classification into categories such as body text, left notes, and footers using a Transformer deep neural network architecture, and finally an aggregation step to compile the lines of a given label in the text. We evaluated the technical performance of the body text extraction algorithm by applying it to a random sample of documents that were annotated. Medical performance was evaluated by examining the extraction of medical concepts of interest from the text in their respective sections. Finally, we tested an end-to-end system on a medical use case of automatic detection of acute infection described in the hospital report. Results:Our algorithm achieved per-line precision, recall, and F1 score of 98.4, 97.0, and 97.7, respectively, for body line extraction. The precision, recall, and F1 score per document for the acute infection detection algorithm were 82.54 (95CI 72.86-91.60), 85.24 (95CI 76.61-93.70), 83.87 (95CI 76, 92-90.08) with exploitation of the results of the advanced body extraction algorithm, respectively. Conclusion:We have developed and validated a system for extracting body text from clinical documents in PDF format by identifying their layout. We were able to demonstrate that this preprocessing allowed us to obtain better performances for a common downstream task, i.e., the extraction of medical concepts in their respective sections, thus proving the interest of this method on a clinical use case.
Development and validation of a natural language processing algorithm to pseudonymize documents in the context of a clinical data warehouse
Mar 23, 2023Abstract:The objective of this study is to address the critical issue of de-identification of clinical reports in order to allow access to data for research purposes, while ensuring patient privacy. The study highlights the difficulties faced in sharing tools and resources in this domain and presents the experience of the Greater Paris University Hospitals (AP-HP) in implementing a systematic pseudonymization of text documents from its Clinical Data Warehouse. We annotated a corpus of clinical documents according to 12 types of identifying entities, and built a hybrid system, merging the results of a deep learning model as well as manual rules. Our results show an overall performance of 0.99 of F1-score. We discuss implementation choices and present experiments to better understand the effort involved in such a task, including dataset size, document types, language models, or rule addition. We share guidelines and code under a 3-Clause BSD license.
Learning structures of the French clinical language:development and validation of word embedding models using 21 million clinical reports from electronic health records
Jul 26, 2022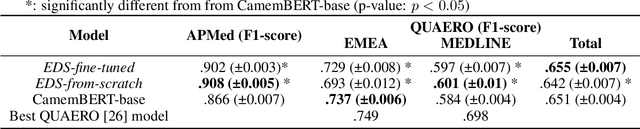
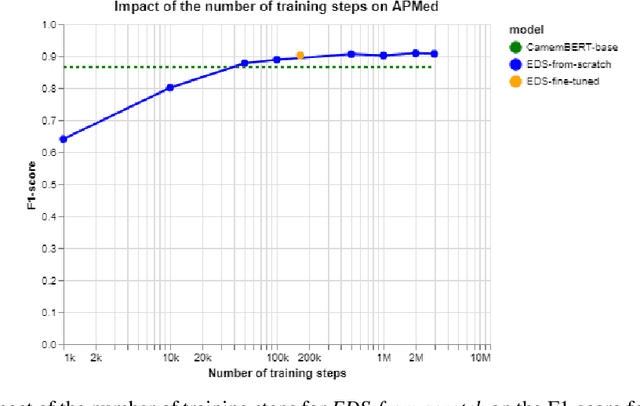
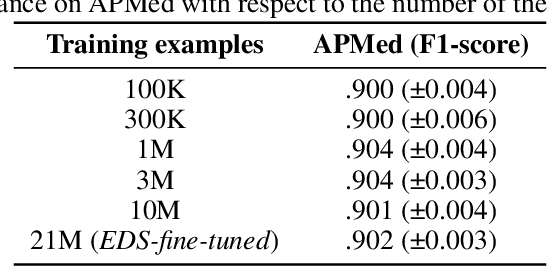
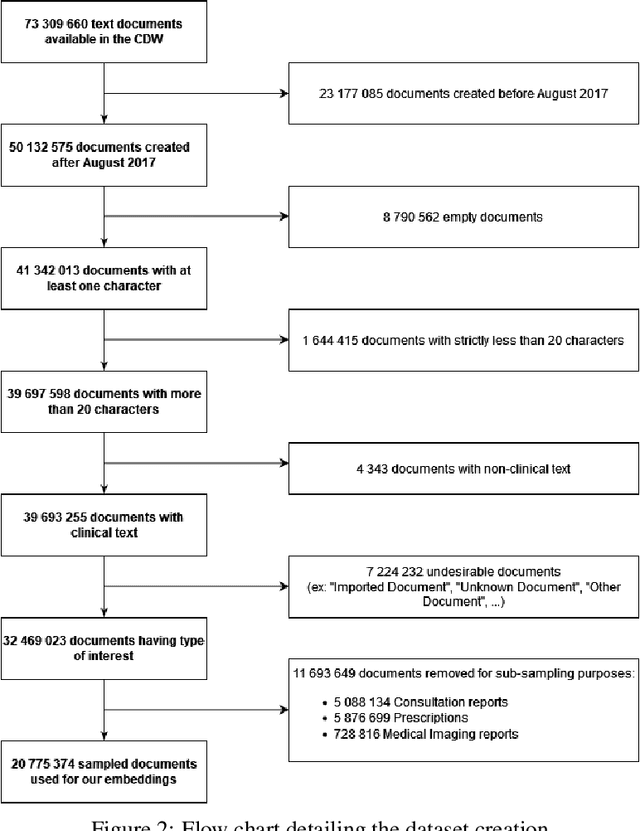
Abstract:Background Clinical studies using real-world data may benefit from exploiting clinical reports, a particularly rich albeit unstructured medium. To that end, natural language processing can extract relevant information. Methods based on transfer learning using pre-trained language models have achieved state-of-the-art results in most NLP applications; however, publicly available models lack exposure to speciality-languages, especially in the medical field. Objective We aimed to evaluate the impact of adapting a language model to French clinical reports on downstream medical NLP tasks. Methods We leveraged a corpus of 21M clinical reports collected from August 2017 to July 2021 at the Greater Paris University Hospitals (APHP) to produce two CamemBERT architectures on speciality language: one retrained from scratch and the other using CamemBERT as its initialisation. We used two French annotated medical datasets to compare our language models to the original CamemBERT network, evaluating the statistical significance of improvement with the Wilcoxon test. Results Our models pretrained on clinical reports increased the average F1-score on APMed (an APHP-specific task) by 3 percentage points to 91%, a statistically significant improvement. They also achieved performance comparable to the original CamemBERT on QUAERO. These results hold true for the fine-tuned and from-scratch versions alike, starting from very few pre-training samples. Conclusions We confirm previous literature showing that adapting generalist pre-train language models such as CamenBERT on speciality corpora improves their performance for downstream clinical NLP tasks. Our results suggest that retraining from scratch does not induce a statistically significant performance gain compared to fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge