Perceval Wajsbürt
Impact of translation on biomedical information extraction from real-life clinical notes
Jun 03, 2023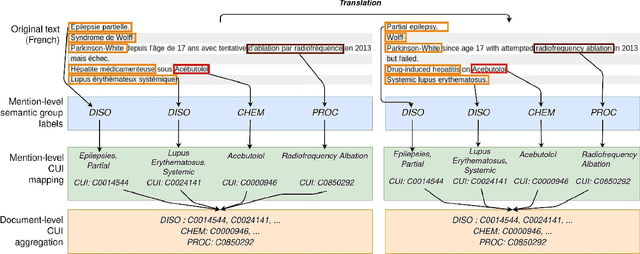
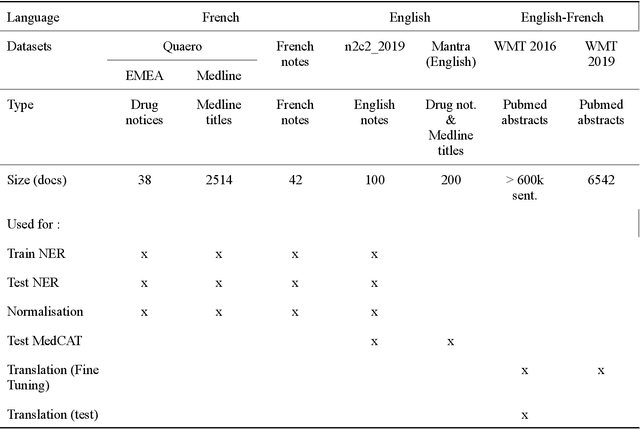
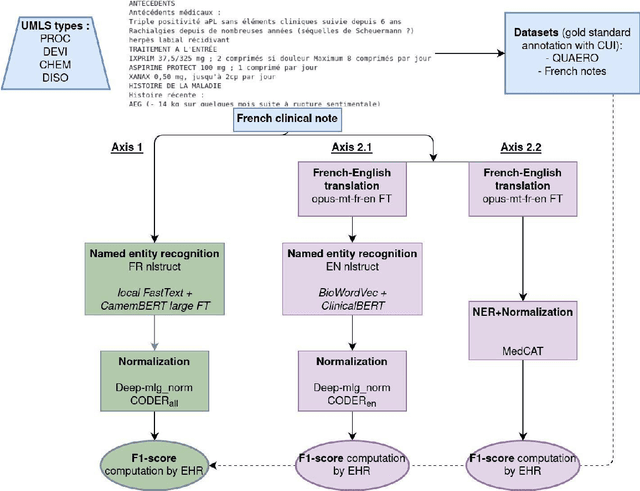
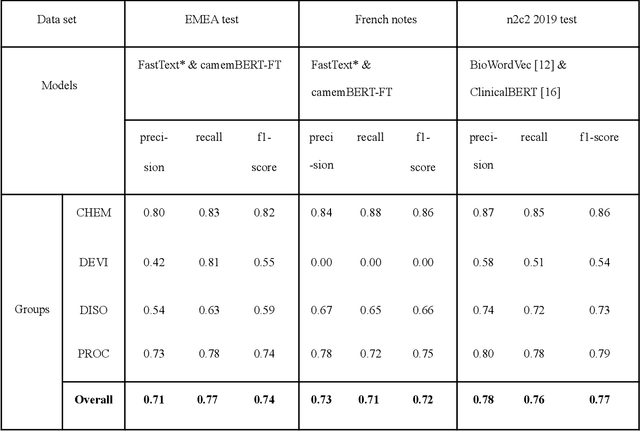
Abstract:The objective of our study is to determine whether using English tools to extract and normalize French medical concepts on translations provides comparable performance to French models trained on a set of annotated French clinical notes. We compare two methods: a method involving French language models and a method involving English language models. For the native French method, the Named Entity Recognition (NER) and normalization steps are performed separately. For the translated English method, after the first translation step, we compare a two-step method and a terminology-oriented method that performs extraction and normalization at the same time. We used French, English and bilingual annotated datasets to evaluate all steps (NER, normalization and translation) of our algorithms. Concerning the results, the native French method performs better than the translated English one with a global f1 score of 0.51 [0.47;0.55] against 0.39 [0.34;0.44] and 0.38 [0.36;0.40] for the two English methods tested. In conclusion, despite the recent improvement of the translation models, there is a significant performance difference between the two approaches in favor of the native French method which is more efficient on French medical texts, even with few annotated documents.
Detecting automatically the layout of clinical documents to enhance the performances of downstream natural language processing
May 23, 2023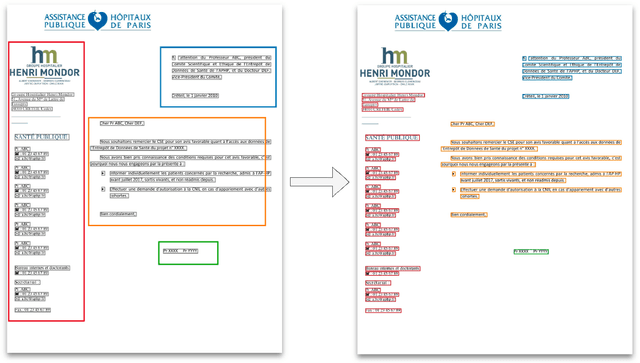


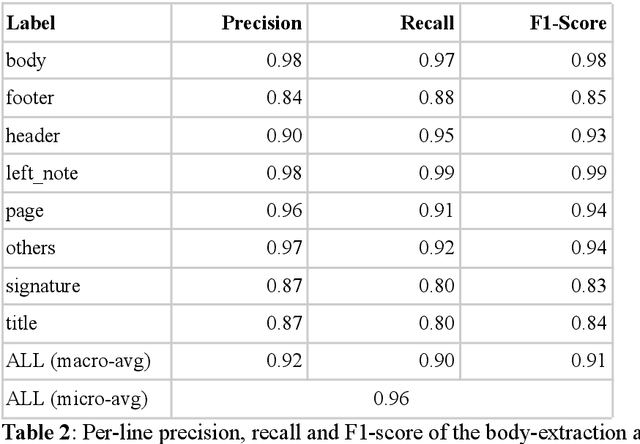
Abstract:Objective:Develop and validate an algorithm for analyzing the layout of PDF clinical documents to improve the performance of downstream natural language processing tasks. Materials and Methods: We designed an algorithm to process clinical PDF documents and extract only clinically relevant text. The algorithm consists of several steps: initial text extraction using a PDF parser, followed by classification into categories such as body text, left notes, and footers using a Transformer deep neural network architecture, and finally an aggregation step to compile the lines of a given label in the text. We evaluated the technical performance of the body text extraction algorithm by applying it to a random sample of documents that were annotated. Medical performance was evaluated by examining the extraction of medical concepts of interest from the text in their respective sections. Finally, we tested an end-to-end system on a medical use case of automatic detection of acute infection described in the hospital report. Results:Our algorithm achieved per-line precision, recall, and F1 score of 98.4, 97.0, and 97.7, respectively, for body line extraction. The precision, recall, and F1 score per document for the acute infection detection algorithm were 82.54 (95CI 72.86-91.60), 85.24 (95CI 76.61-93.70), 83.87 (95CI 76, 92-90.08) with exploitation of the results of the advanced body extraction algorithm, respectively. Conclusion:We have developed and validated a system for extracting body text from clinical documents in PDF format by identifying their layout. We were able to demonstrate that this preprocessing allowed us to obtain better performances for a common downstream task, i.e., the extraction of medical concepts in their respective sections, thus proving the interest of this method on a clinical use case.
Development and validation of a natural language processing algorithm to pseudonymize documents in the context of a clinical data warehouse
Mar 23, 2023Abstract:The objective of this study is to address the critical issue of de-identification of clinical reports in order to allow access to data for research purposes, while ensuring patient privacy. The study highlights the difficulties faced in sharing tools and resources in this domain and presents the experience of the Greater Paris University Hospitals (AP-HP) in implementing a systematic pseudonymization of text documents from its Clinical Data Warehouse. We annotated a corpus of clinical documents according to 12 types of identifying entities, and built a hybrid system, merging the results of a deep learning model as well as manual rules. Our results show an overall performance of 0.99 of F1-score. We discuss implementation choices and present experiments to better understand the effort involved in such a task, including dataset size, document types, language models, or rule addition. We share guidelines and code under a 3-Clause BSD license.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge