Antoine Neuraz
HeKA, UPCité, CRC
Step-by-Step Guidance to Differential Anemia Diagnosis with Real-World Data and Deep Reinforcement Learning
Dec 03, 2024Abstract:Clinical diagnostic guidelines outline the key questions to answer to reach a diagnosis. Inspired by guidelines, we aim to develop a model that learns from electronic health records to determine the optimal sequence of actions for accurate diagnosis. Focusing on anemia and its sub-types, we employ deep reinforcement learning (DRL) algorithms and evaluate their performance on both a synthetic dataset, which is based on expert-defined diagnostic pathways, and a real-world dataset. We investigate the performance of these algorithms across various scenarios. Our experimental results demonstrate that DRL algorithms perform competitively with state-of-the-art methods while offering the significant advantage of progressively generating pathways to the suggested diagnosis, providing a transparent decision-making process that can guide and explain diagnostic reasoning.
Facilitating phenotyping from clinical texts: the medkit library
Aug 30, 2024

Abstract:Phenotyping consists in applying algorithms to identify individuals associated with a specific, potentially complex, trait or condition, typically out of a collection of Electronic Health Records (EHRs). Because a lot of the clinical information of EHRs are lying in texts, phenotyping from text takes an important role in studies that rely on the secondary use of EHRs. However, the heterogeneity and highly specialized aspect of both the content and form of clinical texts makes this task particularly tedious, and is the source of time and cost constraints in observational studies. To facilitate the development, evaluation and reproductibility of phenotyping pipelines, we developed an open-source Python library named medkit. It enables composing data processing pipelines made of easy-to-reuse software bricks, named medkit operations. In addition to the core of the library, we share the operations and pipelines we already developed and invite the phenotyping community for their reuse and enrichment. medkit is available at https://github.com/medkit-lib/medkit
Deep Reinforcement Learning for Personalized Diagnostic Decision Pathways Using Electronic Health Records: A Comparative Study on Anemia and Systemic Lupus Erythematosus
Apr 09, 2024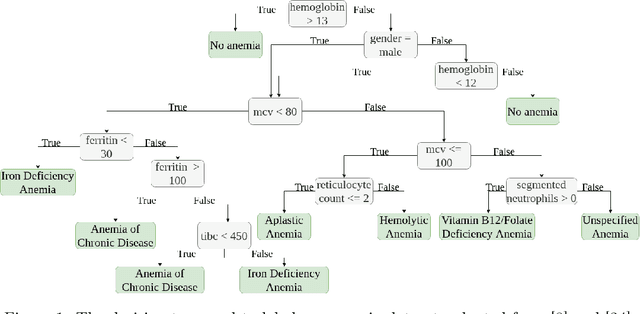

Abstract:Background: Clinical diagnosis is typically reached by following a series of steps recommended by guidelines authored by colleges of experts. Accordingly, guidelines play a crucial role in rationalizing clinical decisions but suffer from limitations as they are built to cover the majority of the population and fail at covering patients with uncommon conditions. Moreover, their updates are long and expensive, making them unsuitable for emerging diseases and practices. Methods: Inspired by guidelines, we formulate the task of diagnosis as a sequential decision-making problem and study the use of Deep Reinforcement Learning (DRL) algorithms to learn the optimal sequence of actions to perform in order to obtain a correct diagnosis from Electronic Health Records (EHRs). We apply DRL on synthetic, but realistic EHRs and develop two clinical use cases: Anemia diagnosis, where the decision pathways follow the schema of a decision tree; and Systemic Lupus Erythematosus (SLE) diagnosis, which follows a weighted criteria score. We particularly evaluate the robustness of our approaches to noisy and missing data since these frequently occur in EHRs. Results: In both use cases, and in the presence of imperfect data, our best DRL algorithms exhibit competitive performance when compared to the traditional classifiers, with the added advantage that they enable the progressive generation of a pathway to the suggested diagnosis which can both guide and explain the decision-making process. Conclusion: DRL offers the opportunity to learn personalized decision pathways to diagnosis. We illustrate with our two use cases their advantages: they generate step-by-step pathways that are self-explanatory; and their correctness is competitive when compared to state-of-the-art approaches.
The Smart Data Extractor, a Clinician Friendly Solution to Accelerate and Improve the Data Collection During Clinical Trials
Aug 31, 2023Abstract:In medical research, the traditional way to collect data, i.e. browsing patient files, has been proven to induce bias, errors, human labor and costs. We propose a semi-automated system able to extract every type of data, including notes. The Smart Data Extractor pre-populates clinic research forms by following rules. We performed a cross-testing experiment to compare semi-automated to manual data collection. 20 target items had to be collected for 79 patients. The average time to complete one form was 6'81'' for manual data collection and 3'22'' with the Smart Data Extractor. There were also more mistakes during manual data collection (163 for the whole cohort) than with the Smart Data Extractor (46 for the whole cohort). We present an easy to use, understandable and agile solution to fill out clinical research forms. It reduces human effort and provides higher quality data, avoiding data re-entry and fatigue induced errors.
Extracting Diagnosis Pathways from Electronic Health Records Using Deep Reinforcement Learning
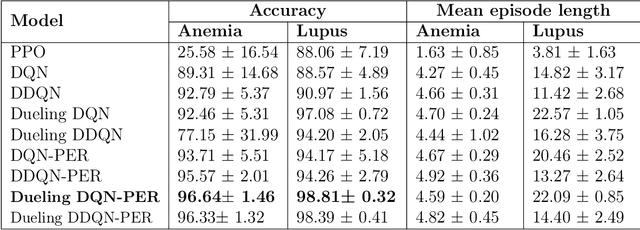
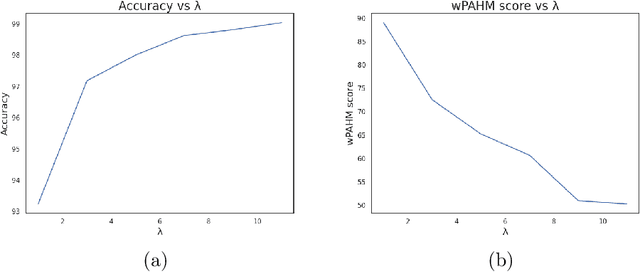
May 10, 2023Abstract:Clinical diagnosis guidelines aim at specifying the steps that may lead to a diagnosis. Guidelines enable rationalizing and normalizing clinical decisions but suffer drawbacks as they are built to cover the majority of the population and may fail in guiding to the right diagnosis for patients with uncommon conditions or multiple pathologies. Moreover, their updates are long and expensive, making them unsuitable to emerging practices. Inspired by guidelines, we formulate the task of diagnosis as a sequential decision-making problem and study the use of Deep Reinforcement Learning (DRL) algorithms trained on Electronic Health Records (EHRs) to learn the optimal sequence of observations to perform in order to obtain a correct diagnosis. Because of the variety of DRL algorithms and of their sensitivity to the context, we considered several approaches and settings that we compared to each other, and to classical classifiers. We experimented on a synthetic but realistic dataset to differentially diagnose anemia and its subtypes and particularly evaluated the robustness of various approaches to noise and missing data as those are frequent in EHRs. Within the DRL algorithms, Dueling DQN with Prioritized Experience Replay, and Dueling Double DQN with Prioritized Experience Replay show the best and most stable performances. In the presence of imperfect data, the DRL algorithms show competitive, but less stable performances when compared to the classifiers (Random Forest and XGBoost); although they enable the progressive generation of a pathway to the suggested diagnosis, which can both guide or explain the decision process.
Learning structures of the French clinical language:development and validation of word embedding models using 21 million clinical reports from electronic health records
Jul 26, 2022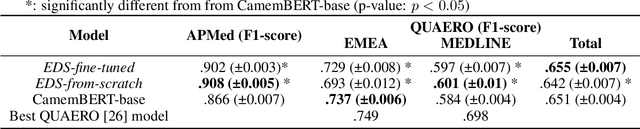
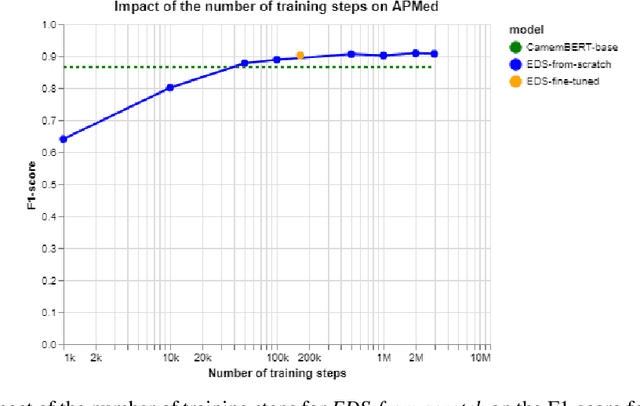
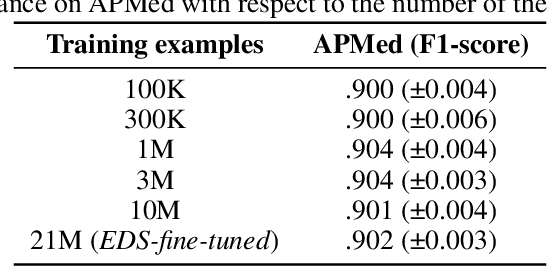
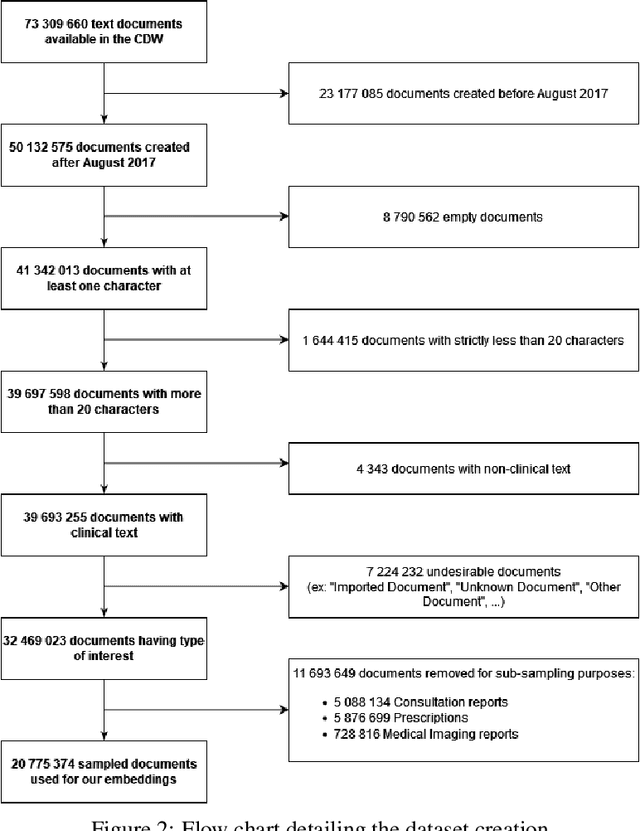
Abstract:Background Clinical studies using real-world data may benefit from exploiting clinical reports, a particularly rich albeit unstructured medium. To that end, natural language processing can extract relevant information. Methods based on transfer learning using pre-trained language models have achieved state-of-the-art results in most NLP applications; however, publicly available models lack exposure to speciality-languages, especially in the medical field. Objective We aimed to evaluate the impact of adapting a language model to French clinical reports on downstream medical NLP tasks. Methods We leveraged a corpus of 21M clinical reports collected from August 2017 to July 2021 at the Greater Paris University Hospitals (APHP) to produce two CamemBERT architectures on speciality language: one retrained from scratch and the other using CamemBERT as its initialisation. We used two French annotated medical datasets to compare our language models to the original CamemBERT network, evaluating the statistical significance of improvement with the Wilcoxon test. Results Our models pretrained on clinical reports increased the average F1-score on APMed (an APHP-specific task) by 3 percentage points to 91%, a statistically significant improvement. They also achieved performance comparable to the original CamemBERT on QUAERO. These results hold true for the fine-tuned and from-scratch versions alike, starting from very few pre-training samples. Conclusions We confirm previous literature showing that adapting generalist pre-train language models such as CamenBERT on speciality corpora improves their performance for downstream clinical NLP tasks. Our results suggest that retraining from scratch does not induce a statistically significant performance gain compared to fine-tuning.
Learning the grammar of prescription: recurrent neural network grammars for medication information extraction in clinical texts
Apr 24, 2020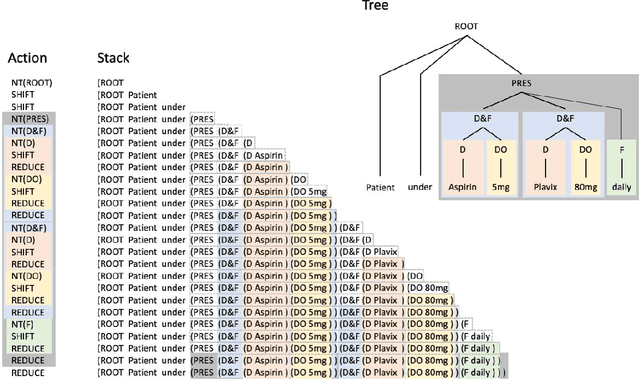
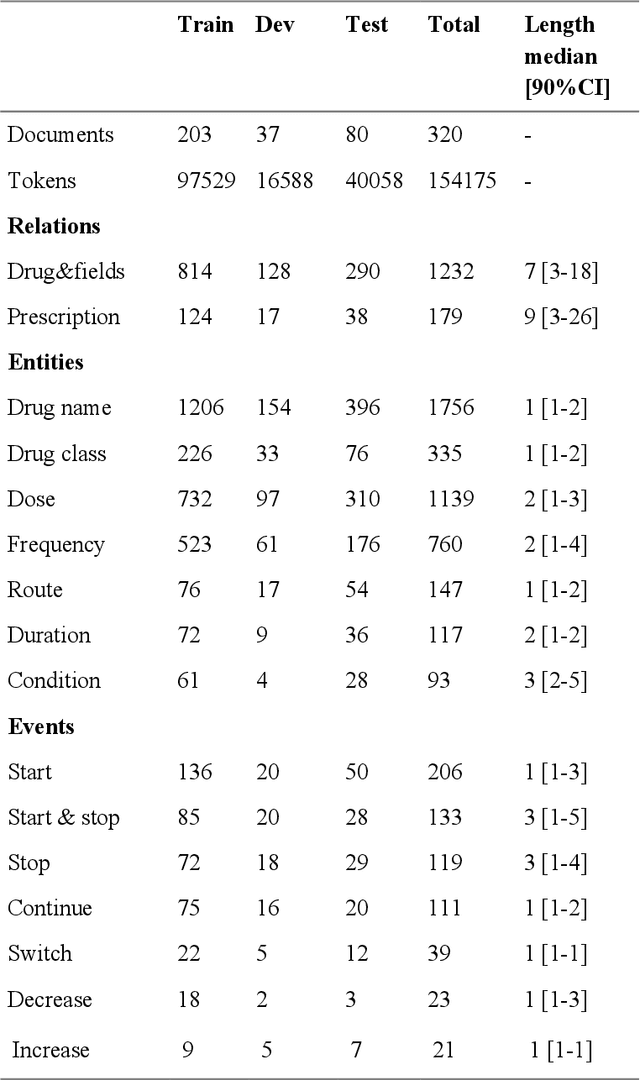
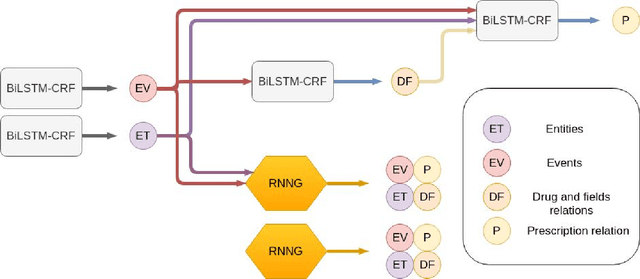
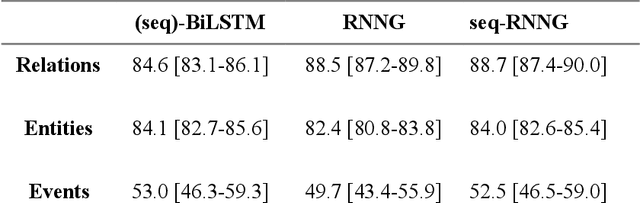
Abstract:In this study, we evaluated the RNNG, a neural top-down transition based parser, for medication information extraction in clinical texts. We evaluated this model on a French clinical corpus. The task was to extract the name of a drug (or class of drug), as well as fields informing its administration: frequency, dosage, duration, condition and route of administration. We compared the RNNG model that jointly identify entities and their relations with separate BiLSTMs models for entities and relations as baselines. We call seq-BiLSTMs the baseline models for relations extraction that takes as extra-input the output of the BiLSTMs for entities. RNNG outperforms seq-BiLSTM for identifying relations, with on average 88.5% [87.2-89.8] versus 84.6 [83.1-86.1] F-measure. However, RNNG is weaker than the baseline BiLSTM on detecting entities, with on average 82.4 [80.8-83.8] versus 84.1 [82.7-85.6] % F- measure. RNNG trained only for detecting relations is weaker than RNNG with the joint modelling objective, 87.4 [85.8-88.8] versus 88.5% [87.2-89.8]. The performance of RNNG on relations can be explained both by the model architecture, which provides shortcut between distant parts of the sentence, and the joint modelling objective which allow the RNNG to learn richer representations. RNNG is efficient for modeling relations between entities in medical texts and its performances are close to those of a BiLSTM for entity detection.
Natural language understanding for task oriented dialog in the biomedical domain in a low resources context
Nov 29, 2018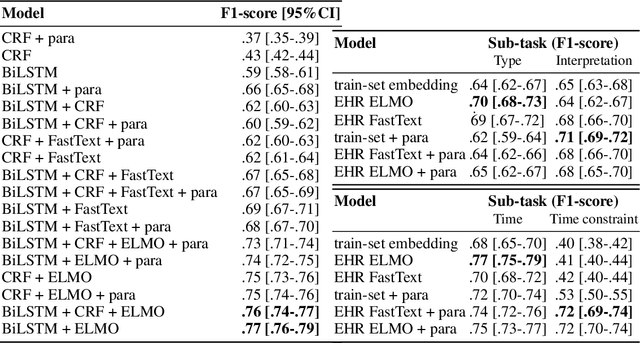
Abstract:In the biomedical domain, the lack of sharable datasets often limit the possibility of developing natural language processing systems, especially dialogue applications and natural language understanding models. To overcome this issue, we explore data generation using templates and terminologies and data augmentation approaches. Namely, we report our experiments using paraphrasing and word representations learned on a large EHR corpus with Fasttext and ELMo, to learn a NLU model without any available dataset. We evaluate on a NLU task of natural language queries in EHRs divided in slot-filling and intent classification sub-tasks. On the slot-filling task, we obtain a F-score of 0.76 with the ELMo representation; and on the classification task, a mean F-score of 0.71. Our results show that this method could be used to develop a baseline system.
A Model-Based Reinforcement Learning Approach for a Rare Disease Diagnostic Task
Nov 25, 2018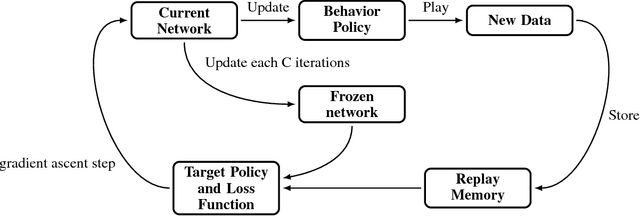
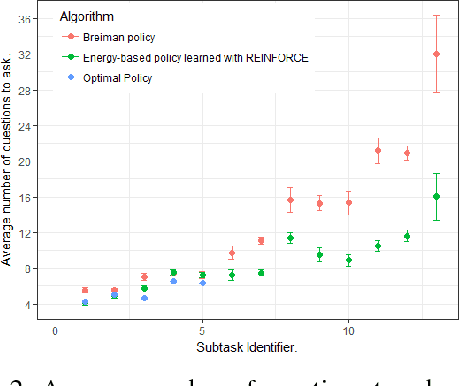
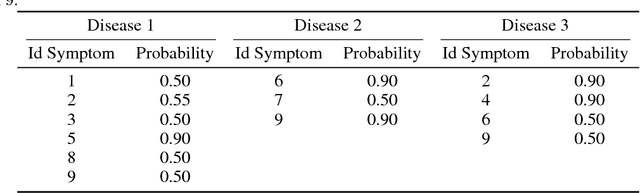
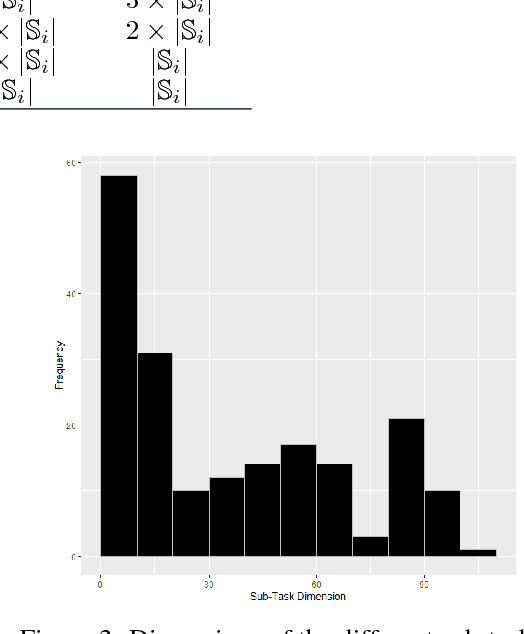
Abstract:In this work, we present our various contributions to the objective of building a decision support tool for the diagnosis of rare diseases. Our goal is to achieve a state of knowledge where the uncertainty about the patient's disease is below a predetermined threshold. We aim to reach such states while minimizing the average number of medical tests to perform. In doing so, we take into account the need, in many medical applications, to avoid, as much as possible, any misdiagnosis. To solve this optimization task, we investigate several reinforcement learning algorithm and make them operable in our high-dimensional and sparse-reward setting. We also present a way to combine expert knowledge, expressed as conditional probabilities, with real clinical data. This is crucial because the scarcity of data in the field of rare diseases prevents any approach based solely on clinical data. Finally we show that it is possible to integrate the ontological information about symptoms while remaining in our probabilistic reasoning. It enables our decision support tool to process information given at different level of precision by the user.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge