Adrien Coulet
Clinical Data Goes MEDS? Let's OWL make sense of it
Jan 07, 2026Abstract:The application of machine learning on healthcare data is often hindered by the lack of standardized and semantically explicit representation, leading to limited interoperability and reproducibility across datasets and experiments. The Medical Event Data Standard (MEDS) addresses these issues by introducing a minimal, event-centric data model designed for reproducible machine-learning workflows from health data. However, MEDS is defined as a data-format specification and does not natively provide integration with the Semantic Web ecosystem. In this article, we introduce MEDS-OWL, a lightweight OWL ontology that provides formal concepts and relations to enable representing MEDS datasets as RDF graphs. Additionally, we implemented meds2rdf, a Python conversion library that transforms MEDS events into RDF graphs, ensuring conformance with the ontology. We demonstrate the approach on a synthetic clinical dataset that describes patient care pathways for ruptured intracranial aneurysms and validate the resulting graph using SHACL constraints. The first release of MEDS-OWL comprises 13 classes, 10 object properties, 20 data properties, and 24 OWL axioms. Combined with meds2rdf, it enables data transformation into FAIR-aligned datasets, provenance-aware publishing, and interoperability of event-based clinical data. By bridging MEDS with the Semantic Web, this work contributes a reusable semantic layer for event-based clinical data and establishes a robust foundation for subsequent graph-based analytics.
Comparing representations of long clinical texts for the task of patient note-identification
Mar 31, 2025



Abstract:In this paper, we address the challenge of patient-note identification, which involves accurately matching an anonymized clinical note to its corresponding patient, represented by a set of related notes. This task has broad applications, including duplicate records detection and patient similarity analysis, which require robust patient-level representations. We explore various embedding methods, including Hierarchical Attention Networks (HAN), three-level Hierarchical Transformer Networks (HTN), LongFormer, and advanced BERT-based models, focusing on their ability to process mediumto-long clinical texts effectively. Additionally, we evaluate different pooling strategies (mean, max, and mean_max) for aggregating wordlevel embeddings into patient-level representations and we examine the impact of sliding windows on model performance. Our results indicate that BERT-based embeddings outperform traditional and hierarchical models, particularly in processing lengthy clinical notes and capturing nuanced patient representations. Among the pooling strategies, mean_max pooling consistently yields the best results, highlighting its ability to capture critical features from clinical notes. Furthermore, the reproduction of our results on both MIMIC dataset and Necker hospital data warehouse illustrates the generalizability of these approaches to real-world applications, emphasizing the importance of both embedding methods and aggregation strategies in optimizing patient-note identification and enhancing patient-level modeling.
Predicting clinical outcomes from patient care pathways represented with temporal knowledge graphs
Feb 28, 2025Abstract:Background: With the increasing availability of healthcare data, predictive modeling finds many applications in the biomedical domain, such as the evaluation of the level of risk for various conditions, which in turn can guide clinical decision making. However, it is unclear how knowledge graph data representations and their embedding, which are competitive in some settings, could be of interest in biomedical predictive modeling. Method: We simulated synthetic but realistic data of patients with intracranial aneurysm and experimented on the task of predicting their clinical outcome. We compared the performance of various classification approaches on tabular data versus a graph-based representation of the same data. Next, we investigated how the adopted schema for representing first individual data and second temporal data impacts predictive performances. Results: Our study illustrates that in our case, a graph representation and Graph Convolutional Network (GCN) embeddings reach the best performance for a predictive task from observational data. We emphasize the importance of the adopted schema and of the consideration of literal values in the representation of individual data. Our study also moderates the relative impact of various time encoding on GCN performance.
Efficient extraction of medication information from clinical notes: an evaluation in two languages
Feb 05, 2025



Abstract:Objective: To evaluate the accuracy, computational cost and portability of a new Natural Language Processing (NLP) method for extracting medication information from clinical narratives. Materials and Methods: We propose an original transformer-based architecture for the extraction of entities and their relations pertaining to patients' medication regimen. First, we used this approach to train and evaluate a model on French clinical notes, using a newly annotated corpus from H\^opitaux Universitaires de Strasbourg. Second, the portability of the approach was assessed by conducting an evaluation on clinical documents in English from the 2018 n2c2 shared task. Information extraction accuracy and computational cost were assessed by comparison with an available method using transformers. Results: The proposed architecture achieves on the task of relation extraction itself performance that are competitive with the state-of-the-art on both French and English (F-measures 0.82 and 0.96 vs 0.81 and 0.95), but reduce the computational cost by 10. End-to-end (Named Entity recognition and Relation Extraction) F1 performance is 0.69 and 0.82 for French and English corpus. Discussion: While an existing system developed for English notes was deployed in a French hospital setting with reasonable effort, we found that an alternative architecture offered end-to-end drug information extraction with comparable extraction performance and lower computational impact for both French and English clinical text processing, respectively. Conclusion: The proposed architecture can be used to extract medication information from clinical text with high performance and low computational cost and consequently suits with usually limited hospital IT resources
Step-by-Step Guidance to Differential Anemia Diagnosis with Real-World Data and Deep Reinforcement Learning
Dec 03, 2024Abstract:Clinical diagnostic guidelines outline the key questions to answer to reach a diagnosis. Inspired by guidelines, we aim to develop a model that learns from electronic health records to determine the optimal sequence of actions for accurate diagnosis. Focusing on anemia and its sub-types, we employ deep reinforcement learning (DRL) algorithms and evaluate their performance on both a synthetic dataset, which is based on expert-defined diagnostic pathways, and a real-world dataset. We investigate the performance of these algorithms across various scenarios. Our experimental results demonstrate that DRL algorithms perform competitively with state-of-the-art methods while offering the significant advantage of progressively generating pathways to the suggested diagnosis, providing a transparent decision-making process that can guide and explain diagnostic reasoning.
Facilitating phenotyping from clinical texts: the medkit library
Aug 30, 2024

Abstract:Phenotyping consists in applying algorithms to identify individuals associated with a specific, potentially complex, trait or condition, typically out of a collection of Electronic Health Records (EHRs). Because a lot of the clinical information of EHRs are lying in texts, phenotyping from text takes an important role in studies that rely on the secondary use of EHRs. However, the heterogeneity and highly specialized aspect of both the content and form of clinical texts makes this task particularly tedious, and is the source of time and cost constraints in observational studies. To facilitate the development, evaluation and reproductibility of phenotyping pipelines, we developed an open-source Python library named medkit. It enables composing data processing pipelines made of easy-to-reuse software bricks, named medkit operations. In addition to the core of the library, we share the operations and pipelines we already developed and invite the phenotyping community for their reuse and enrichment. medkit is available at https://github.com/medkit-lib/medkit
Deep Reinforcement Learning for Personalized Diagnostic Decision Pathways Using Electronic Health Records: A Comparative Study on Anemia and Systemic Lupus Erythematosus
Apr 09, 2024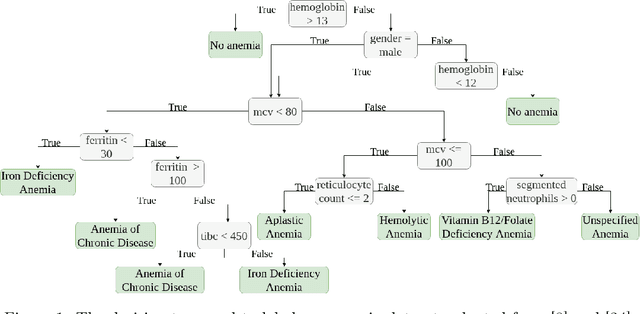
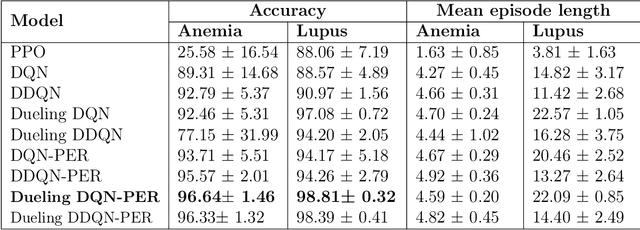

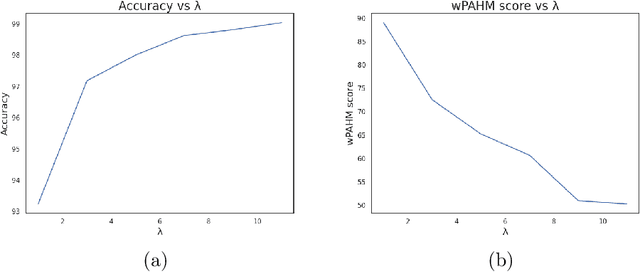
Abstract:Background: Clinical diagnosis is typically reached by following a series of steps recommended by guidelines authored by colleges of experts. Accordingly, guidelines play a crucial role in rationalizing clinical decisions but suffer from limitations as they are built to cover the majority of the population and fail at covering patients with uncommon conditions. Moreover, their updates are long and expensive, making them unsuitable for emerging diseases and practices. Methods: Inspired by guidelines, we formulate the task of diagnosis as a sequential decision-making problem and study the use of Deep Reinforcement Learning (DRL) algorithms to learn the optimal sequence of actions to perform in order to obtain a correct diagnosis from Electronic Health Records (EHRs). We apply DRL on synthetic, but realistic EHRs and develop two clinical use cases: Anemia diagnosis, where the decision pathways follow the schema of a decision tree; and Systemic Lupus Erythematosus (SLE) diagnosis, which follows a weighted criteria score. We particularly evaluate the robustness of our approaches to noisy and missing data since these frequently occur in EHRs. Results: In both use cases, and in the presence of imperfect data, our best DRL algorithms exhibit competitive performance when compared to the traditional classifiers, with the added advantage that they enable the progressive generation of a pathway to the suggested diagnosis which can both guide and explain the decision-making process. Conclusion: DRL offers the opportunity to learn personalized decision pathways to diagnosis. We illustrate with our two use cases their advantages: they generate step-by-step pathways that are self-explanatory; and their correctness is competitive when compared to state-of-the-art approaches.
Extracting Diagnosis Pathways from Electronic Health Records Using Deep Reinforcement Learning
May 10, 2023Abstract:Clinical diagnosis guidelines aim at specifying the steps that may lead to a diagnosis. Guidelines enable rationalizing and normalizing clinical decisions but suffer drawbacks as they are built to cover the majority of the population and may fail in guiding to the right diagnosis for patients with uncommon conditions or multiple pathologies. Moreover, their updates are long and expensive, making them unsuitable to emerging practices. Inspired by guidelines, we formulate the task of diagnosis as a sequential decision-making problem and study the use of Deep Reinforcement Learning (DRL) algorithms trained on Electronic Health Records (EHRs) to learn the optimal sequence of observations to perform in order to obtain a correct diagnosis. Because of the variety of DRL algorithms and of their sensitivity to the context, we considered several approaches and settings that we compared to each other, and to classical classifiers. We experimented on a synthetic but realistic dataset to differentially diagnose anemia and its subtypes and particularly evaluated the robustness of various approaches to noise and missing data as those are frequent in EHRs. Within the DRL algorithms, Dueling DQN with Prioritized Experience Replay, and Dueling Double DQN with Prioritized Experience Replay show the best and most stable performances. In the presence of imperfect data, the DRL algorithms show competitive, but less stable performances when compared to the classifiers (Random Forest and XGBoost); although they enable the progressive generation of a pathway to the suggested diagnosis, which can both guide or explain the decision process.
Investigating ADR mechanisms with knowledge graph mining and explainable AI
Dec 16, 2020
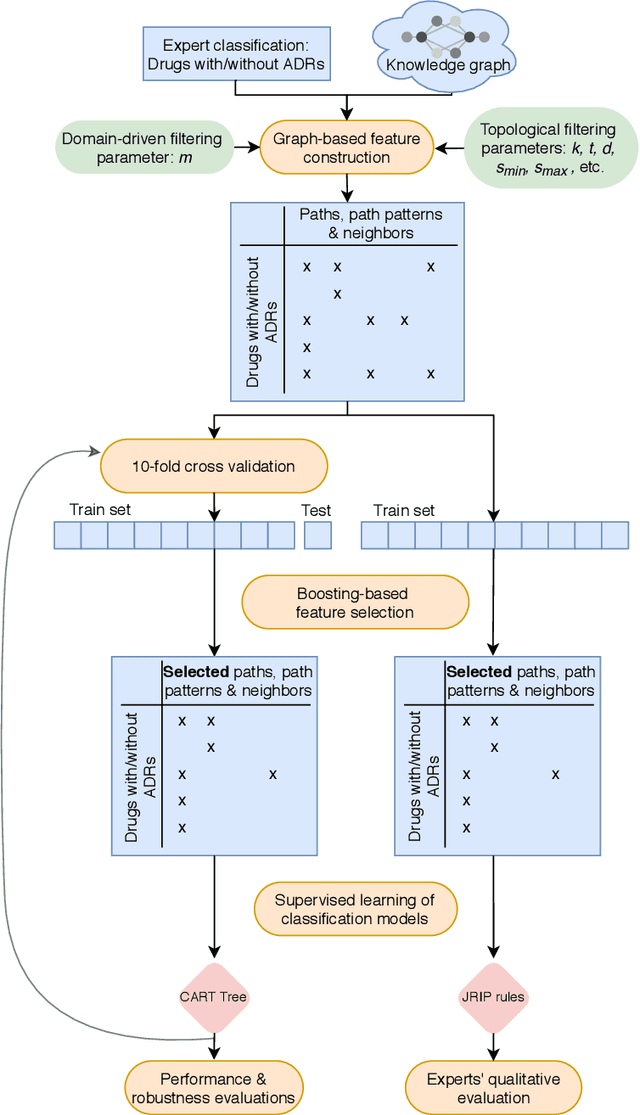


Abstract:Adverse Drug Reactions (ADRs) are characterized within randomized clinical trials and postmarketing pharmacovigilance, but their molecular mechanism remains unknown in most cases. Aside from clinical trials, many elements of knowledge about drug ingredients are available in open-access knowledge graphs. In addition, drug classifications that label drugs as either causative or not for several ADRs, have been established. We propose to mine knowledge graphs for identifying biomolecular features that may enable reproducing automatically expert classifications that distinguish drug causative or not for a given type of ADR. In an explainable AI perspective, we explore simple classification techniques such as Decision Trees and Classification Rules because they provide human-readable models, which explain the classification itself, but may also provide elements of explanation for molecular mechanisms behind ADRs. In summary, we mine a knowledge graph for features; we train classifiers at distinguishing, drugs associated or not with ADRs; we isolate features that are both efficient in reproducing expert classifications and interpretable by experts (i.e., Gene Ontology terms, drug targets, or pathway names); and we manually evaluate how they may be explanatory. Extracted features reproduce with a good fidelity classifications of drugs causative or not for DILI and SCAR. Experts fully agreed that 73% and 38% of the most discriminative features are possibly explanatory for DILI and SCAR, respectively; and partially agreed (2/3) for 90% and 77% of them. Knowledge graphs provide diverse features to enable simple and explainable models to distinguish between drugs that are causative or not for ADRs. In addition to explaining classifications, most discriminative features appear to be good candidates for investigating ADR mechanisms further.
Experiments on transfer learning architectures for biomedical relation extraction
Nov 24, 2020
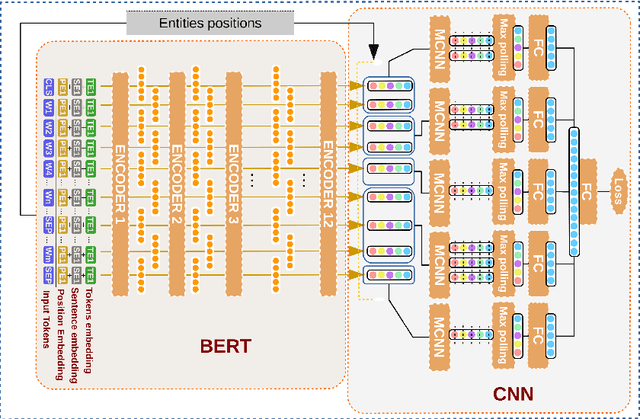
Abstract:Relation extraction (RE) consists in identifying and structuring automatically relations of interest from texts. Recently, BERT improved the top performances for several NLP tasks, including RE. However, the best way to use BERT, within a machine learning architecture, and within a transfer learning strategy is still an open question since it is highly dependent on each specific task and domain. Here, we explore various BERT-based architectures and transfer learning strategies (i.e., frozen or fine-tuned) for the task of biomedical RE on two corpora. Among tested architectures and strategies, our *BERT-segMCNN with finetuning reaches performances higher than the state-of-the-art on the two corpora (1.73 % and 32.77 % absolute improvement on ChemProt and PGxCorpus corpora respectively). More generally, our experiments illustrate the expected interest of fine-tuning with BERT, but also the unexplored advantage of using structural information (with sentence segmentation), in addition to the context classically leveraged by BERT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge