Malika Smaïl-Tabbone
ORPAILLEUR
Investigating ADR mechanisms with knowledge graph mining and explainable AI
Dec 16, 2020
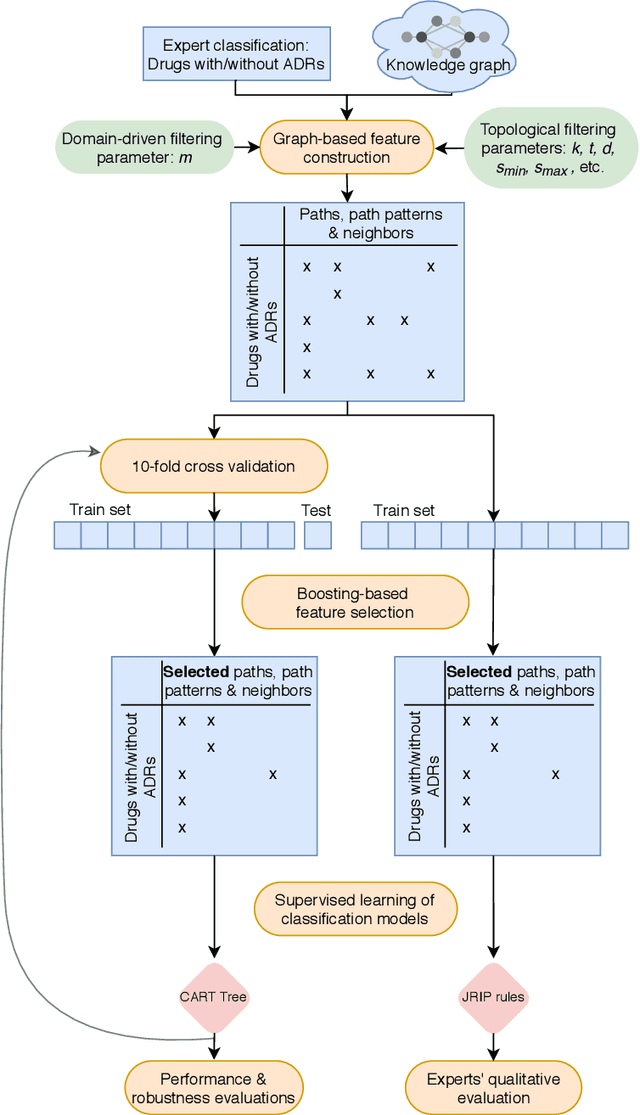


Abstract:Adverse Drug Reactions (ADRs) are characterized within randomized clinical trials and postmarketing pharmacovigilance, but their molecular mechanism remains unknown in most cases. Aside from clinical trials, many elements of knowledge about drug ingredients are available in open-access knowledge graphs. In addition, drug classifications that label drugs as either causative or not for several ADRs, have been established. We propose to mine knowledge graphs for identifying biomolecular features that may enable reproducing automatically expert classifications that distinguish drug causative or not for a given type of ADR. In an explainable AI perspective, we explore simple classification techniques such as Decision Trees and Classification Rules because they provide human-readable models, which explain the classification itself, but may also provide elements of explanation for molecular mechanisms behind ADRs. In summary, we mine a knowledge graph for features; we train classifiers at distinguishing, drugs associated or not with ADRs; we isolate features that are both efficient in reproducing expert classifications and interpretable by experts (i.e., Gene Ontology terms, drug targets, or pathway names); and we manually evaluate how they may be explanatory. Extracted features reproduce with a good fidelity classifications of drugs causative or not for DILI and SCAR. Experts fully agreed that 73% and 38% of the most discriminative features are possibly explanatory for DILI and SCAR, respectively; and partially agreed (2/3) for 90% and 77% of them. Knowledge graphs provide diverse features to enable simple and explainable models to distinguish between drugs that are causative or not for ADRs. In addition to explaining classifications, most discriminative features appear to be good candidates for investigating ADR mechanisms further.
Tackling scalability issues in mining path patterns from knowledge graphs: a preliminary study
Aug 07, 2020



Abstract:Features mined from knowledge graphs are widely used within multiple knowledge discovery tasks such as classification or fact-checking. Here, we consider a given set of vertices, called seed vertices, and focus on mining their associated neighboring vertices, paths, and, more generally, path patterns that involve classes of ontologies linked with knowledge graphs. Due to the combinatorial nature and the increasing size of real-world knowledge graphs, the task of mining these patterns immediately entails scalability issues. In this paper, we address these issues by proposing a pattern mining approach that relies on a set of constraints (e.g., support or degree thresholds) and the monotonicity property. As our motivation comes from the mining of real-world knowledge graphs, we illustrate our approach with PGxLOD, a biomedical knowledge graph.
Neighborhood-Based Label Propagation in Large Protein Graphs
Aug 09, 2017
Abstract:Understanding protein function is one of the keys to understanding life at the molecular level. It is also important in several scenarios including human disease and drug discovery. In this age of rapid and affordable biological sequencing, the number of sequences accumulating in databases is rising with an increasing rate. This presents many challenges for biologists and computer scientists alike. In order to make sense of this huge quantity of data, these sequences should be annotated with functional properties. UniProtKB consists of two components: i) the UniProtKB/Swiss-Prot database containing protein sequences with reliable information manually reviewed by expert bio-curators and ii) the UniProtKB/TrEMBL database that is used for storing and processing the unknown sequences. Hence, for all proteins we have available the sequence along with few more information such as the taxon and some structural domains. Pairwise similarity can be defined and computed on proteins based on such attributes. Other important attributes, while present for proteins in Swiss-Prot, are often missing for proteins in TrEMBL, such as their function and cellular localization. The enormous number of protein sequences now in TrEMBL calls for rapid procedures to annotate them automatically. In this work, we present DistNBLP, a novel Distributed Neighborhood-Based Label Propagation approach for large-scale annotation of proteins. To do this, the functional annotations of reviewed proteins are used to predict those of non-reviewed proteins using label propagation on a graph representation of the protein database. DistNBLP is built on top of the "akka" toolkit for building resilient distributed message-driven applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge