Pierre Monnin
WIMMICS
Eat your own KR: a KR-based approach to index Semantic Web Endpoints and Knowledge Graphs
Aug 12, 2025Abstract:Over the last decade, knowledge graphs have multiplied, grown, and evolved on the World Wide Web, and the advent of new standards, vocabularies, and application domains has accelerated this trend. IndeGx is a framework leveraging an extensible base of rules to index the content of KGs and the capacities of their SPARQL endpoints. In this article, we show how knowledge representation (KR) and reasoning methods and techniques can be used in a reflexive manner to index and characterize existing knowledge graphs (KG) with respect to their usage of KR methods and techniques. We extended IndeGx with a fully ontology-oriented modeling and processing approach to do so. Using SPARQL rules and an OWL RL ontology of the indexing domain, IndeGx can now build and reason over an index of the contents and characteristics of an open collection of public knowledge graphs. Our extension of the framework relies on a declarative representation of procedural knowledge and collaborative environments (e.g., GitHub) to provide an agile, customizable, and expressive KR approach for building and maintaining such an index of knowledge graphs in the wild. In doing so, we help anyone answer the question of what knowledge is out there in the world wild Semantic Web in general, and we also help our community monitor which KR research results are used in practice. In particular, this article provides a snapshot of the state of the Semantic Web regarding supported standard languages, ontology usage, and diverse quality evaluations by applying this method to a collection of over 300 open knowledge graph endpoints.
KGPrune: a Web Application to Extract Subgraphs of Interest from Wikidata with Analogical Pruning
Aug 26, 2024Abstract:Knowledge graphs (KGs) have become ubiquitous publicly available knowledge sources, and are nowadays covering an ever increasing array of domains. However, not all knowledge represented is useful or pertaining when considering a new application or specific task. Also, due to their increasing size, handling large KGs in their entirety entails scalability issues. These two aspects asks for efficient methods to extract subgraphs of interest from existing KGs. To this aim, we introduce KGPrune, a Web Application that, given seed entities of interest and properties to traverse, extracts their neighboring subgraphs from Wikidata. To avoid topical drift, KGPrune relies on a frugal pruning algorithm based on analogical reasoning to only keep relevant neighbors while pruning irrelevant ones. The interest of KGPrune is illustrated by two concrete applications, namely, bootstrapping an enterprise KG and extracting knowledge related to looted artworks.
Beyond Transduction: A Survey on Inductive, Few Shot, and Zero Shot Link Prediction in Knowledge Graphs
Dec 08, 2023
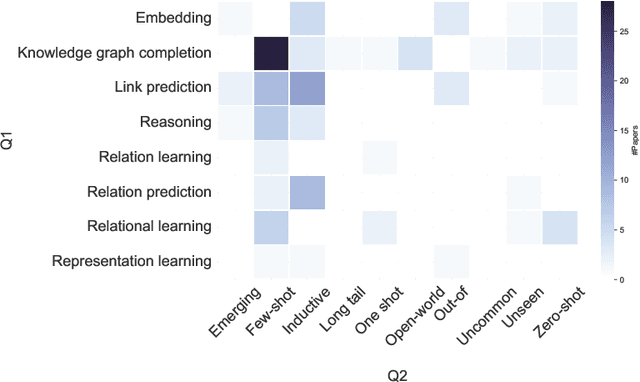
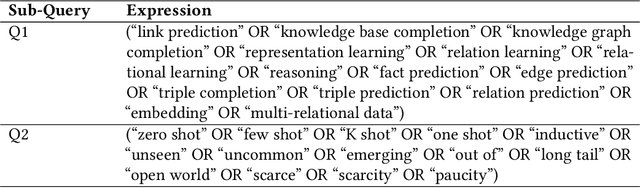
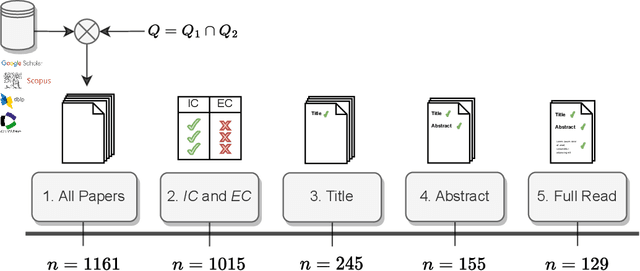
Abstract:Knowledge graphs (KGs) comprise entities interconnected by relations of different semantic meanings. KGs are being used in a wide range of applications. However, they inherently suffer from incompleteness, i.e. entities or facts about entities are missing. Consequently, a larger body of works focuses on the completion of missing information in KGs, which is commonly referred to as link prediction (LP). This task has traditionally and extensively been studied in the transductive setting, where all entities and relations in the testing set are observed during training. Recently, several works have tackled the LP task under more challenging settings, where entities and relations in the test set may be unobserved during training, or appear in only a few facts. These works are known as inductive, few-shot, and zero-shot link prediction. In this work, we conduct a systematic review of existing works in this area. A thorough analysis leads us to point out the undesirable existence of diverging terminologies and task definitions for the aforementioned settings, which further limits the possibility of comparison between recent works. We consequently aim at dissecting each setting thoroughly, attempting to reveal its intrinsic characteristics. A unifying nomenclature is ultimately proposed to refer to each of them in a simple and consistent manner.
Knowledge Graphs for the Life Sciences: Recent Developments, Challenges and Opportunities
Oct 06, 2023Abstract:The term life sciences refers to the disciplines that study living organisms and life processes, and include chemistry, biology, medicine, and a range of other related disciplines. Research efforts in life sciences are heavily data-driven, as they produce and consume vast amounts of scientific data, much of which is intrinsically relational and graph-structured. The volume of data and the complexity of scientific concepts and relations referred to therein promote the application of advanced knowledge-driven technologies for managing and interpreting data, with the ultimate aim to advance scientific discovery. In this survey and position paper, we discuss recent developments and advances in the use of graph-based technologies in life sciences and set out a vision for how these technologies will impact these fields into the future. We focus on three broad topics: the construction and management of Knowledge Graphs (KGs), the use of KGs and associated technologies in the discovery of new knowledge, and the use of KGs in artificial intelligence applications to support explanations (explainable AI). We select a few exemplary use cases for each topic, discuss the challenges and open research questions within these topics, and conclude with a perspective and outlook that summarizes the overarching challenges and their potential solutions as a guide for future research.
PyGraft: Configurable Generation of Schemas and Knowledge Graphs at Your Fingertips
Sep 07, 2023Abstract:Knowledge graphs (KGs) have emerged as a prominent data representation and management paradigm. Being usually underpinned by a schema (e.g. an ontology), KGs capture not only factual information but also contextual knowledge. In some tasks, a few KGs established themselves as standard benchmarks. However, recent works outline that relying on a limited collection of datasets is not sufficient to assess the generalization capability of an approach. In some data-sensitive fields such as education or medicine, access to public datasets is even more limited. To remedy the aforementioned issues, we release PyGraft, a Python-based tool that generates highly customized, domain-agnostic schemas and knowledge graphs. The synthesized schemas encompass various RDFS and OWL constructs, while the synthesized KGs emulate the characteristics and scale of real-world KGs. Logical consistency of the generated resources is ultimately ensured by running a description logic (DL) reasoner. By providing a way of generating both a schema and KG in a single pipeline, PyGraft's aim is to empower the generation of a more diverse array of KGs for benchmarking novel approaches in areas such as graph-based machine learning (ML), or more generally KG processing. In graph-based ML in particular, this should foster a more holistic evaluation of model performance and generalization capability, thereby going beyond the limited collection of available benchmarks. PyGraft is available at: https://github.com/nicolas-hbt/pygraft.
Relevant Entity Selection: Knowledge Graph Bootstrapping via Zero-Shot Analogical Pruning
Jun 28, 2023



Abstract:Knowledge Graph Construction (KGC) can be seen as an iterative process starting from a high quality nucleus that is refined by knowledge extraction approaches in a virtuous loop. Such a nucleus can be obtained from knowledge existing in an open KG like Wikidata. However, due to the size of such generic KGs, integrating them as a whole may entail irrelevant content and scalability issues. We propose an analogy-based approach that starts from seed entities of interest in a generic KG, and keeps or prunes their neighboring entities. We evaluate our approach on Wikidata through two manually labeled datasets that contain either domain-homogeneous or -heterogeneous seed entities. We empirically show that our analogy-based approach outperforms LSTM, Random Forest, SVM, and MLP, with a drastically lower number of parameters. We also evaluate its generalization potential in a transfer learning setting. These results advocate for the further integration of analogy-based inference in tasks related to the KG lifecycle.
Schema First! Learn Versatile Knowledge Graph Embeddings by Capturing Semantics with MASCHInE
Jun 06, 2023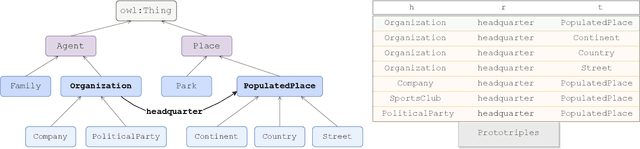
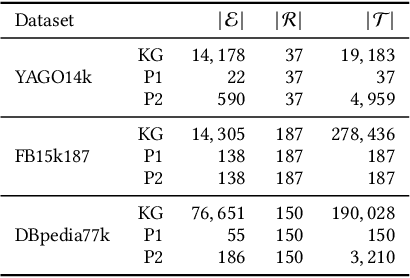
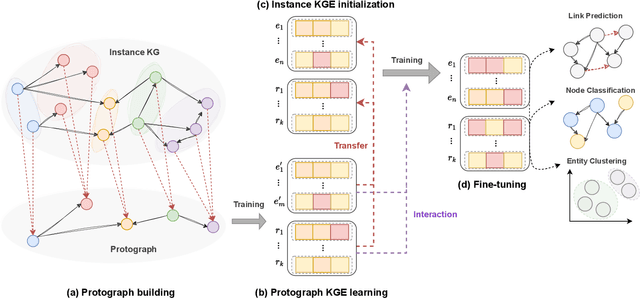
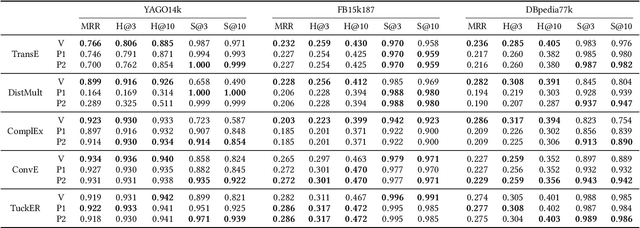
Abstract:Knowledge graph embedding models (KGEMs) have gained considerable traction in recent years. These models learn a vector representation of knowledge graph entities and relations, a.k.a. knowledge graph embeddings (KGEs). Learning versatile KGEs is desirable as it makes them useful for a broad range of tasks. However, KGEMs are usually trained for a specific task, which makes their embeddings task-dependent. In parallel, the widespread assumption that KGEMs actually create a semantic representation of the underlying entities and relations (e.g., project similar entities closer than dissimilar ones) has been challenged. In this work, we design heuristics for generating protographs -- small, modified versions of a KG that leverage schema-based information. The learnt protograph-based embeddings are meant to encapsulate the semantics of a KG, and can be leveraged in learning KGEs that, in turn, also better capture semantics. Extensive experiments on various evaluation benchmarks demonstrate the soundness of this approach, which we call Modular and Agnostic SCHema-based Integration of protograph Embeddings (MASCHInE). In particular, MASCHInE helps produce more versatile KGEs that yield substantially better performance for entity clustering and node classification tasks. For link prediction, using MASCHInE has little impact on rank-based performance but increases the number of semantically valid predictions.
Enhancing Knowledge Graph Embedding Models with Semantic-driven Loss Functions
Mar 09, 2023Abstract:Knowledge graph embedding models (KGEMs) are used for various tasks related to knowledge graphs (KGs), including link prediction. They are trained with loss functions that are computed considering a batch of scored triples and their corresponding labels. Traditional approaches consider the label of a triple to be either true or false. However, recent works suggest that all negative triples should not be valued equally. In line with this recent assumption, we posit that semantically valid negative triples might be high-quality negative triples. As such, loss functions should treat them differently from semantically invalid negative ones. To this aim, we propose semantic-driven versions for the three main loss functions for link prediction. In particular, we treat the scores of negative triples differently by injecting background knowledge about relation domains and ranges into the loss functions. In an extensive and controlled experimental setting, we show that the proposed loss functions systematically provide satisfying results on three public benchmark KGs underpinned with different schemas, which demonstrates both the generality and superiority of our proposed approach. In fact, the proposed loss functions do (1) lead to better MRR and Hits@$10$ values, (2) drive KGEMs towards better semantic awareness. This highlights that semantic information globally improves KGEMs, and thus should be incorporated into loss functions. Domains and ranges of relations being largely available in schema-defined KGs, this makes our approach both beneficial and widely usable in practice.
Sem@$K$: Is my knowledge graph embedding model semantic-aware?
Jan 13, 2023



Abstract:Using knowledge graph embedding models (KGEMs) is a popular approach for predicting links in knowledge graphs (KGs). Traditionally, the performance of KGEMs for link prediction is assessed using rank-based metrics, which evaluate their ability to give high scores to ground-truth entities. However, the literature claims that the KGEM evaluation procedure would benefit from adding supplementary dimensions to assess. That is why, in this paper, we extend our previously introduced metric Sem@$K$ that measures the capability of models to predict valid entities w.r.t. domain and range constrains. In particular, we consider a broad range of KGs and take their respective characteristics into account to propose different versions of Sem@$K$. We also perform an extensive study of KGEM semantic awareness. Our experiments show that Sem@$K$ provides a new perspective on KGEM quality. Its joint analysis with rank-based metrics offer different conclusions on the predictive power of models. Regarding Sem@$K$, some KGEMs are inherently better than others, but this semantic superiority is not indicative of their performance w.r.t. rank-based metrics. In this work, we generalize conclusions about the relative performance of KGEMs w.r.t. rank-based and semantic-oriented metrics at the level of families of models. The joint analysis of the aforementioned metrics gives more insight into the peculiarities of each model. This work paves the way for a more comprehensive evaluation of KGEM adequacy for specific downstream tasks.
Investigating ADR mechanisms with knowledge graph mining and explainable AI
Dec 16, 2020
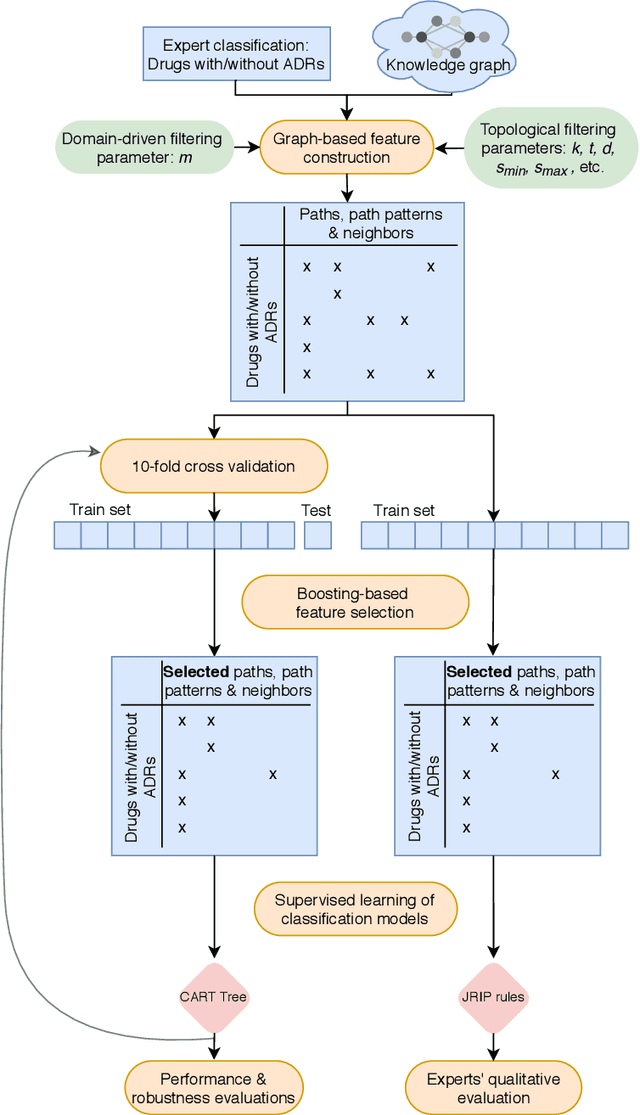


Abstract:Adverse Drug Reactions (ADRs) are characterized within randomized clinical trials and postmarketing pharmacovigilance, but their molecular mechanism remains unknown in most cases. Aside from clinical trials, many elements of knowledge about drug ingredients are available in open-access knowledge graphs. In addition, drug classifications that label drugs as either causative or not for several ADRs, have been established. We propose to mine knowledge graphs for identifying biomolecular features that may enable reproducing automatically expert classifications that distinguish drug causative or not for a given type of ADR. In an explainable AI perspective, we explore simple classification techniques such as Decision Trees and Classification Rules because they provide human-readable models, which explain the classification itself, but may also provide elements of explanation for molecular mechanisms behind ADRs. In summary, we mine a knowledge graph for features; we train classifiers at distinguishing, drugs associated or not with ADRs; we isolate features that are both efficient in reproducing expert classifications and interpretable by experts (i.e., Gene Ontology terms, drug targets, or pathway names); and we manually evaluate how they may be explanatory. Extracted features reproduce with a good fidelity classifications of drugs causative or not for DILI and SCAR. Experts fully agreed that 73% and 38% of the most discriminative features are possibly explanatory for DILI and SCAR, respectively; and partially agreed (2/3) for 90% and 77% of them. Knowledge graphs provide diverse features to enable simple and explainable models to distinguish between drugs that are causative or not for ADRs. In addition to explaining classifications, most discriminative features appear to be good candidates for investigating ADR mechanisms further.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge