Valentina Tamma
Evaluating the Fitness of Ontologies for the Task of Question Generation
Apr 08, 2025Abstract:Ontology-based question generation is an important application of semantic-aware systems that enables the creation of large question banks for diverse learning environments. The effectiveness of these systems, both in terms of the calibre and cognitive difficulty of the resulting questions, depends heavily on the quality and modelling approach of the underlying ontologies, making it crucial to assess their fitness for this task. To date, there has been no comprehensive investigation into the specific ontology aspects or characteristics that affect the question generation process. Therefore, this paper proposes a set of requirements and task-specific metrics for evaluating the fitness of ontologies for question generation tasks in pedagogical settings. Using the ROMEO methodology, a structured framework for deriving task-specific metrics, an expert-based approach is employed to assess the performance of various ontologies in Automatic Question Generation (AQG) tasks, which is then evaluated over a set of ontologies. Our results demonstrate that ontology characteristics significantly impact the effectiveness of question generation, with different ontologies exhibiting varying performance levels. This highlights the importance of assessing ontology quality with respect to AQG tasks.
A Pattern to Align Them All: Integrating Different Modalities to Define Multi-Modal Entities
Oct 17, 2024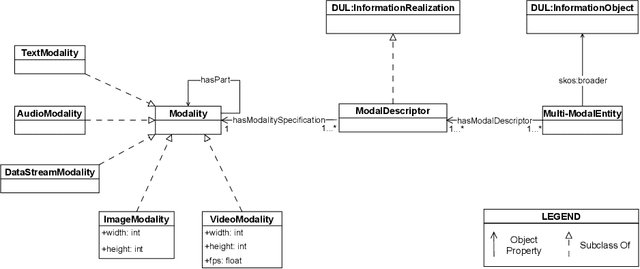

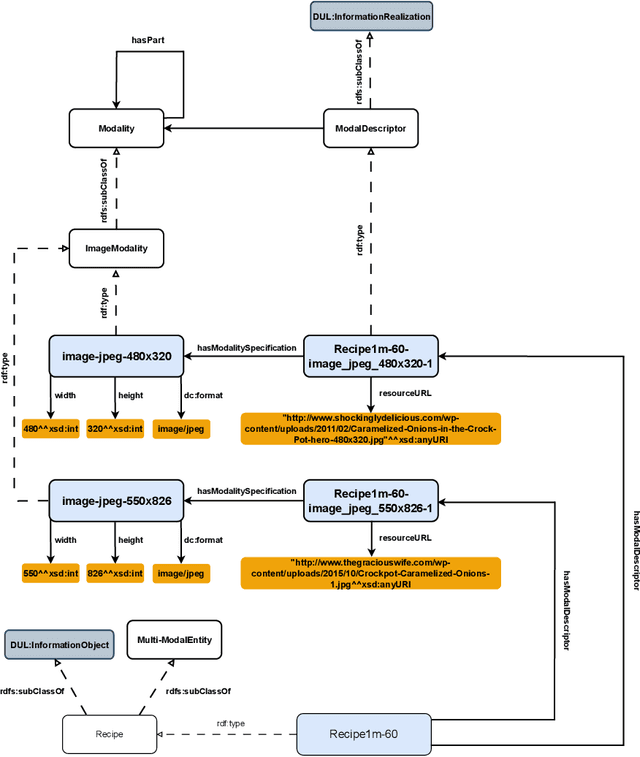
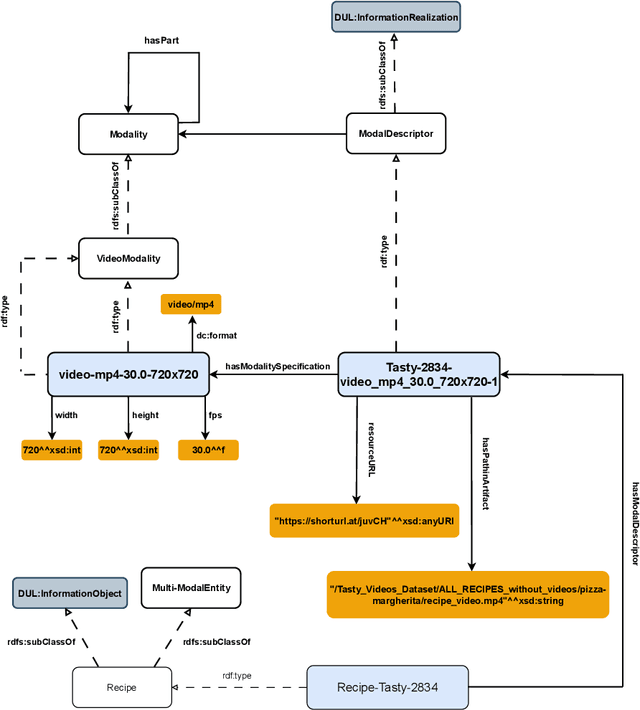
Abstract:The ability to reason with and integrate different sensory inputs is the foundation underpinning human intelligence and it is the reason for the growing interest in modelling multi-modal information within Knowledge Graphs. Multi-Modal Knowledge Graphs extend traditional Knowledge Graphs by associating an entity with its possible modal representations, including text, images, audio, and videos, all of which are used to convey the semantics of the entity. Despite the increasing attention that Multi-Modal Knowledge Graphs have received, there is a lack of consensus about the definitions and modelling of modalities, whose definition is often determined by application domains. In this paper, we propose a novel ontology design pattern that captures the separation of concerns between an entity (and the information it conveys), whose semantics can have different manifestations across different media, and its realisation in terms of a physical information entity. By introducing this abstract model, we aim to facilitate the harmonisation and integration of different existing multi-modal ontologies which is crucial for many intelligent applications across different domains spanning from medicine to digital humanities.
An Experiment in Retrofitting Competency Questions for Existing Ontologies
Nov 09, 2023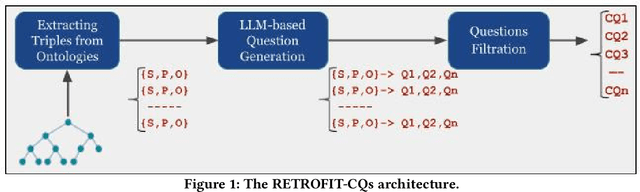
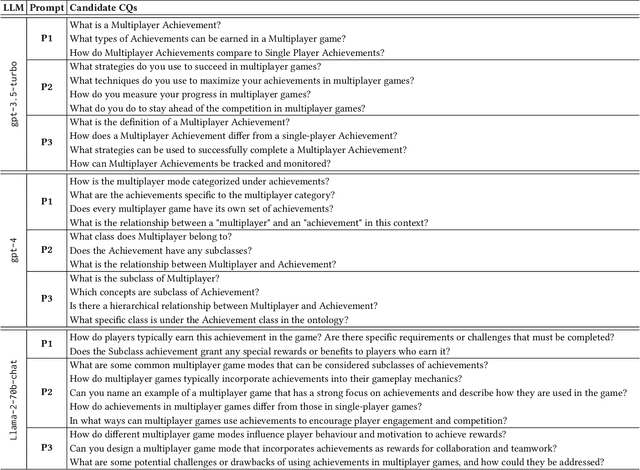
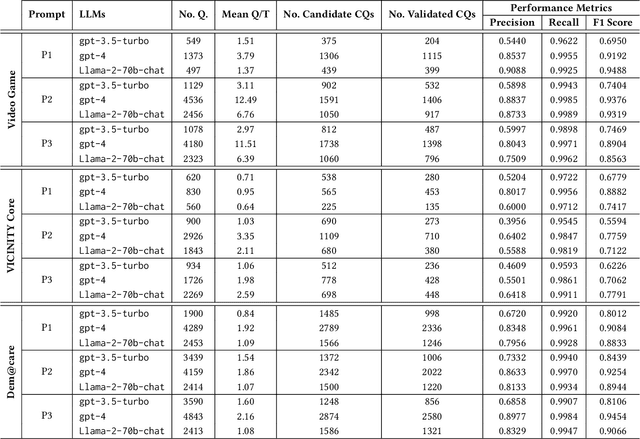
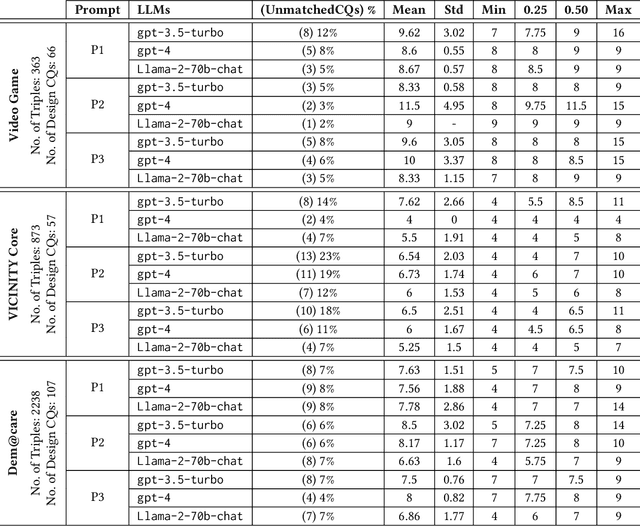
Abstract:Competency Questions (CQs) are a form of ontology functional requirements expressed as natural language questions. Inspecting CQs together with the axioms in an ontology provides critical insights into the intended scope and applicability of the ontology. CQs also underpin a number of tasks in the development of ontologies e.g. ontology reuse, ontology testing, requirement specification, and the definition of patterns that implement such requirements. Although CQs are integral to the majority of ontology engineering methodologies, the practice of publishing CQs alongside the ontological artefacts is not widely observed by the community. In this context, we present an experiment in retrofitting CQs from existing ontologies. We propose RETROFIT-CQs, a method to extract candidate CQs directly from ontologies using Generative AI. In the paper we present the pipeline that facilitates the extraction of CQs by leveraging Large Language Models (LLMs) and we discuss its application to a number of existing ontologies.
Knowledge Graphs for the Life Sciences: Recent Developments, Challenges and Opportunities
Oct 06, 2023Abstract:The term life sciences refers to the disciplines that study living organisms and life processes, and include chemistry, biology, medicine, and a range of other related disciplines. Research efforts in life sciences are heavily data-driven, as they produce and consume vast amounts of scientific data, much of which is intrinsically relational and graph-structured. The volume of data and the complexity of scientific concepts and relations referred to therein promote the application of advanced knowledge-driven technologies for managing and interpreting data, with the ultimate aim to advance scientific discovery. In this survey and position paper, we discuss recent developments and advances in the use of graph-based technologies in life sciences and set out a vision for how these technologies will impact these fields into the future. We focus on three broad topics: the construction and management of Knowledge Graphs (KGs), the use of KGs and associated technologies in the discovery of new knowledge, and the use of KGs in artificial intelligence applications to support explanations (explainable AI). We select a few exemplary use cases for each topic, discuss the challenges and open research questions within these topics, and conclude with a perspective and outlook that summarizes the overarching challenges and their potential solutions as a guide for future research.
OntoMerger: An Ontology Integration Library for Deduplicating and Connecting Knowledge Graph Nodes
Jun 05, 2022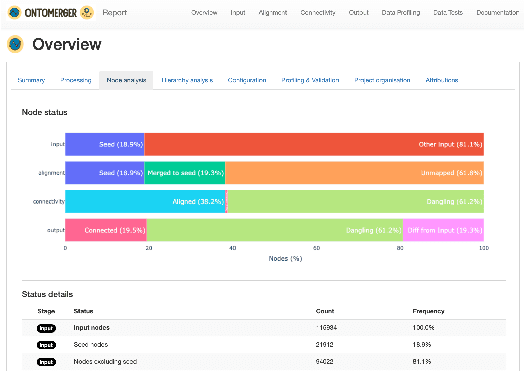
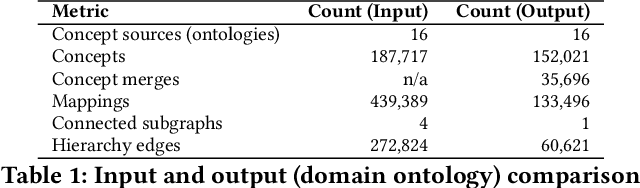
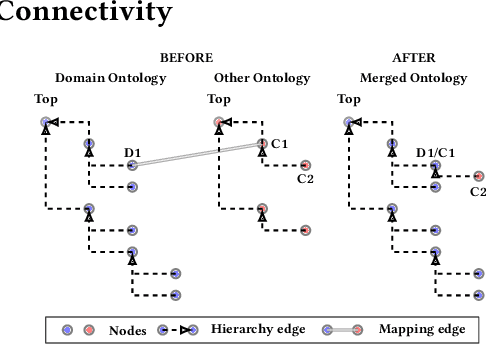

Abstract:Duplication of nodes is a common problem encountered when building knowledge graphs (KGs) from heterogeneous datasets, where it is crucial to be able to merge nodes having the same meaning. OntoMerger is a Python ontology integration library whose functionality is to deduplicate KG nodes. Our approach takes a set of KG nodes, mappings and disconnected hierarchies and generates a set of merged nodes together with a connected hierarchy. In addition, the library provides analytic and data testing functionalities that can be used to fine-tune the inputs, further reducing duplication, and to increase connectivity of the output graph. OntoMerger can be applied to a wide variety of ontologies and KGs. In this paper we introduce OntoMerger and illustrate its functionality on a real-world biomedical KG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge