Vanessa López
WaveGAS: Waveform Relaxation for Scaling Graph Neural Networks
Feb 27, 2025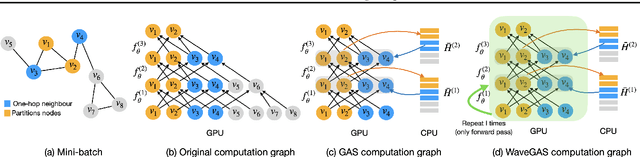
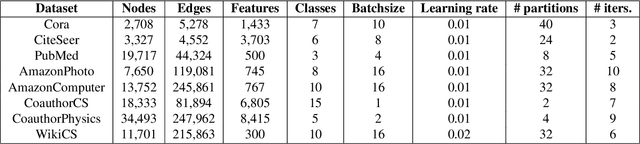
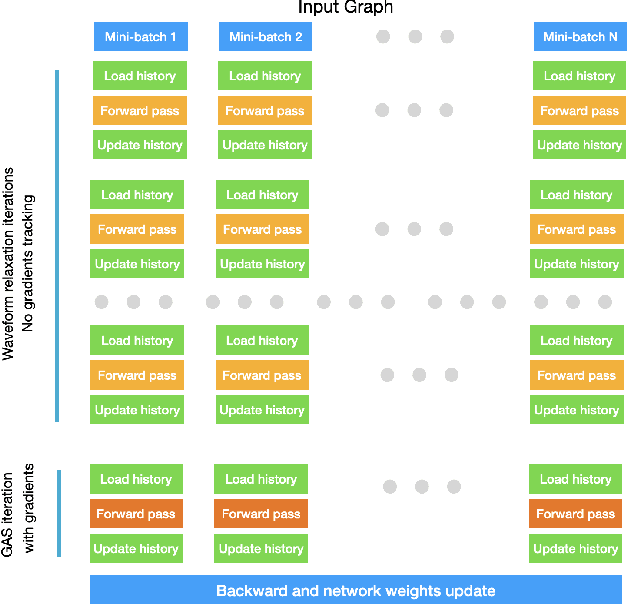
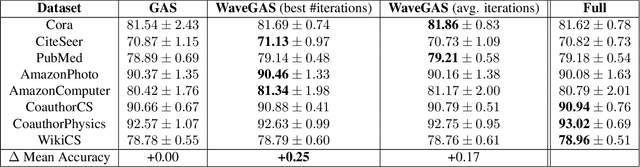
Abstract:With the ever-growing size of real-world graphs, numerous techniques to overcome resource limitations when training Graph Neural Networks (GNNs) have been developed. One such approach, GNNAutoScale (GAS), uses graph partitioning to enable training under constrained GPU memory. GAS also stores historical embedding vectors, which are retrieved from one-hop neighbors in other partitions, ensuring critical information is captured across partition boundaries. The historical embeddings which come from the previous training iteration are stale compared to the GAS estimated embeddings, resulting in approximation errors of the training algorithm. Furthermore, these errors accumulate over multiple layers, leading to suboptimal node embeddings. To address this shortcoming, we propose two enhancements: first, WaveGAS, inspired by waveform relaxation, performs multiple forward passes within GAS before the backward pass, refining the approximation of historical embeddings and gradients to improve accuracy; second, a gradient-tracking method that stores and utilizes more accurate historical gradients during training. Empirical results show that WaveGAS enhances GAS and achieves better accuracy, even outperforming methods that train on full graphs, thanks to its robust estimation of node embeddings.
Description Boosting for Zero-Shot Entity and Relation Classification
Jun 04, 2024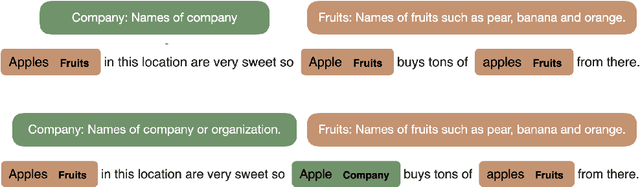

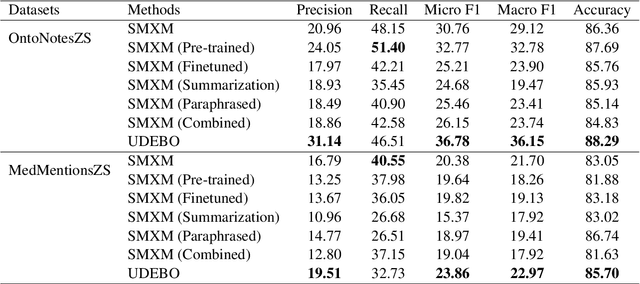
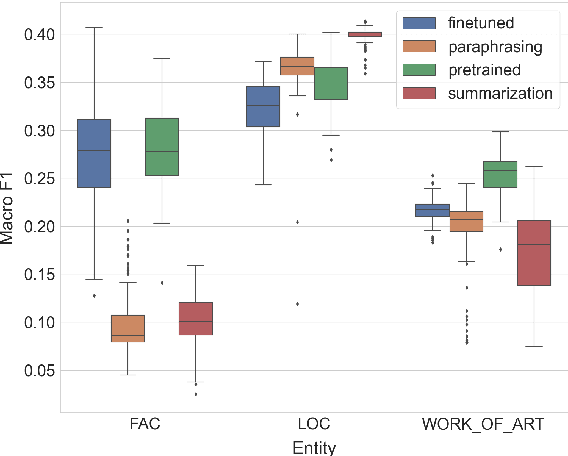
Abstract:Zero-shot entity and relation classification models leverage available external information of unseen classes -- e.g., textual descriptions -- to annotate input text data. Thanks to the minimum data requirement, Zero-Shot Learning (ZSL) methods have high value in practice, especially in applications where labeled data is scarce. Even though recent research in ZSL has demonstrated significant results, our analysis reveals that those methods are sensitive to provided textual descriptions of entities (or relations). Even a minor modification of descriptions can lead to a change in the decision boundary between entity (or relation) classes. In this paper, we formally define the problem of identifying effective descriptions for zero shot inference. We propose a strategy for generating variations of an initial description, a heuristic for ranking them and an ensemble method capable of boosting the predictions of zero-shot models through description enhancement. Empirical results on four different entity and relation classification datasets show that our proposed method outperform existing approaches and achieve new SOTA results on these datasets under the ZSL settings. The source code of the proposed solutions and the evaluation framework are open-sourced.
Knowledge Graphs for the Life Sciences: Recent Developments, Challenges and Opportunities
Oct 06, 2023Abstract:The term life sciences refers to the disciplines that study living organisms and life processes, and include chemistry, biology, medicine, and a range of other related disciplines. Research efforts in life sciences are heavily data-driven, as they produce and consume vast amounts of scientific data, much of which is intrinsically relational and graph-structured. The volume of data and the complexity of scientific concepts and relations referred to therein promote the application of advanced knowledge-driven technologies for managing and interpreting data, with the ultimate aim to advance scientific discovery. In this survey and position paper, we discuss recent developments and advances in the use of graph-based technologies in life sciences and set out a vision for how these technologies will impact these fields into the future. We focus on three broad topics: the construction and management of Knowledge Graphs (KGs), the use of KGs and associated technologies in the discovery of new knowledge, and the use of KGs in artificial intelligence applications to support explanations (explainable AI). We select a few exemplary use cases for each topic, discuss the challenges and open research questions within these topics, and conclude with a perspective and outlook that summarizes the overarching challenges and their potential solutions as a guide for future research.
Zshot: An Open-source Framework for Zero-Shot Named Entity Recognition and Relation Extraction
Jul 25, 2023Abstract:The Zero-Shot Learning (ZSL) task pertains to the identification of entities or relations in texts that were not seen during training. ZSL has emerged as a critical research area due to the scarcity of labeled data in specific domains, and its applications have grown significantly in recent years. With the advent of large pretrained language models, several novel methods have been proposed, resulting in substantial improvements in ZSL performance. There is a growing demand, both in the research community and industry, for a comprehensive ZSL framework that facilitates the development and accessibility of the latest methods and pretrained models.In this study, we propose a novel ZSL framework called Zshot that aims to address the aforementioned challenges. Our primary objective is to provide a platform that allows researchers to compare different state-of-the-art ZSL methods with standard benchmark datasets. Additionally, we have designed our framework to support the industry with readily available APIs for production under the standard SpaCy NLP pipeline. Our API is extendible and evaluable, moreover, we include numerous enhancements such as boosting the accuracy with pipeline ensembling and visualization utilities available as a SpaCy extension.
* Accepted at ACL 2023
Otter-Knowledge: benchmarks of multimodal knowledge graph representation learning from different sources for drug discovery
Jun 23, 2023Abstract:Recent research in representation learning utilizes large databases of proteins or molecules to acquire knowledge of drug and protein structures through unsupervised learning techniques. These pre-trained representations have proven to significantly enhance the accuracy of subsequent tasks, such as predicting the affinity between drugs and target proteins. In this study, we demonstrate that by incorporating knowledge graphs from diverse sources and modalities into the sequences or SMILES representation, we can further enrich the representation and achieve state-of-the-art results on established benchmark datasets. We provide preprocessed and integrated data obtained from 7 public sources, which encompass over 30M triples. Additionally, we make available the pre-trained models based on this data, along with the reported outcomes of their performance on three widely-used benchmark datasets for drug-target binding affinity prediction found in the Therapeutic Data Commons (TDC) benchmarks. Additionally, we make the source code for training models on benchmark datasets publicly available. Our objective in releasing these pre-trained models, accompanied by clean data for model pretraining and benchmark results, is to encourage research in knowledge-enhanced representation learning.
Ensembling Graph Predictions for AMR Parsing
Oct 18, 2021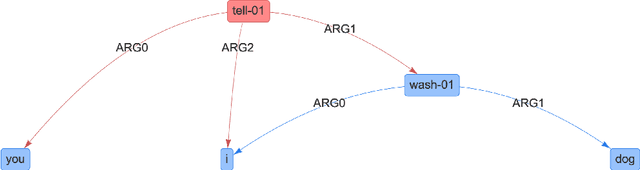
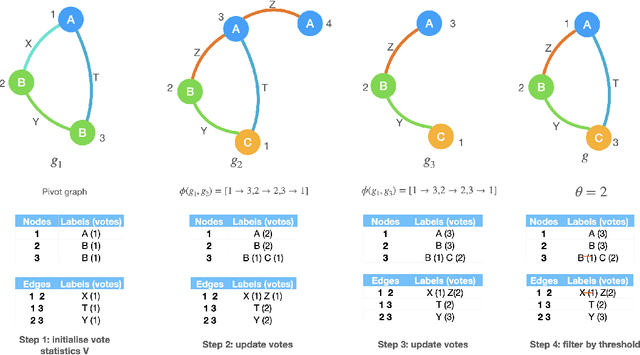
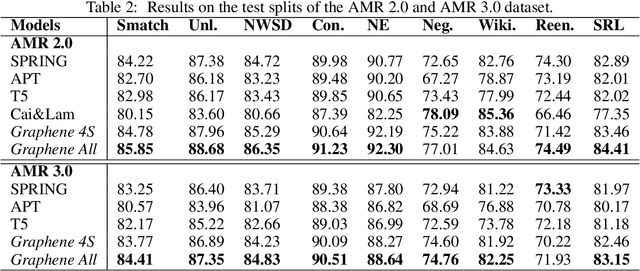
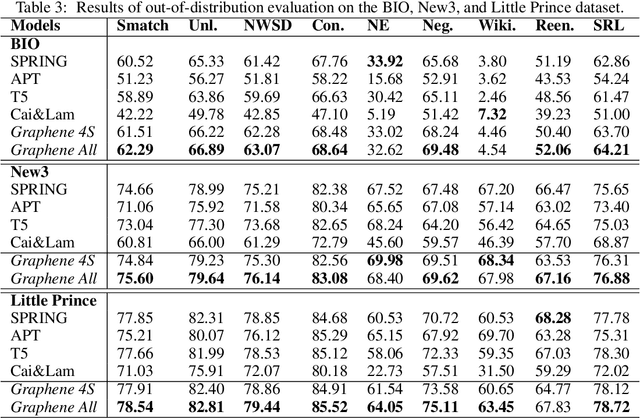
Abstract:In many machine learning tasks, models are trained to predict structure data such as graphs. For example, in natural language processing, it is very common to parse texts into dependency trees or abstract meaning representation (AMR) graphs. On the other hand, ensemble methods combine predictions from multiple models to create a new one that is more robust and accurate than individual predictions. In the literature, there are many ensembling techniques proposed for classification or regression problems, however, ensemble graph prediction has not been studied thoroughly. In this work, we formalize this problem as mining the largest graph that is the most supported by a collection of graph predictions. As the problem is NP-Hard, we propose an efficient heuristic algorithm to approximate the optimal solution. To validate our approach, we carried out experiments in AMR parsing problems. The experimental results demonstrate that the proposed approach can combine the strength of state-of-the-art AMR parsers to create new predictions that are more accurate than any individual models in five standard benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge