Janna Hastings
Chemical classification program synthesis using generative artificial intelligence
May 24, 2025Abstract:Accurately classifying chemical structures is essential for cheminformatics and bioinformatics, including tasks such as identifying bioactive compounds of interest, screening molecules for toxicity to humans, finding non-organic compounds with desirable material properties, or organizing large chemical libraries for drug discovery or environmental monitoring. However, manual classification is labor-intensive and difficult to scale to large chemical databases. Existing automated approaches either rely on manually constructed classification rules, or the use of deep learning methods that lack explainability. This work presents an approach that uses generative artificial intelligence to automatically write chemical classifier programs for classes in the Chemical Entities of Biological Interest (ChEBI) database. These programs can be used for efficient deterministic run-time classification of SMILES structures, with natural language explanations. The programs themselves constitute an explainable computable ontological model of chemical class nomenclature, which we call the ChEBI Chemical Class Program Ontology (C3PO). We validated our approach against the ChEBI database, and compared our results against state of the art deep learning models. We also demonstrate the use of C3PO to classify out-of-distribution examples taken from metabolomics repositories and natural product databases. We also demonstrate the potential use of our approach to find systematic classification errors in existing chemical databases, and show how an ensemble artificial intelligence approach combining generated ontologies, automated literature search, and multimodal vision models can be used to pinpoint potential errors requiring expert validation
Cross-Care: Assessing the Healthcare Implications of Pre-training Data on Language Model Bias
May 09, 2024



Abstract:Large language models (LLMs) are increasingly essential in processing natural languages, yet their application is frequently compromised by biases and inaccuracies originating in their training data. In this study, we introduce Cross-Care, the first benchmark framework dedicated to assessing biases and real world knowledge in LLMs, specifically focusing on the representation of disease prevalence across diverse demographic groups. We systematically evaluate how demographic biases embedded in pre-training corpora like $ThePile$ influence the outputs of LLMs. We expose and quantify discrepancies by juxtaposing these biases against actual disease prevalences in various U.S. demographic groups. Our results highlight substantial misalignment between LLM representation of disease prevalence and real disease prevalence rates across demographic subgroups, indicating a pronounced risk of bias propagation and a lack of real-world grounding for medical applications of LLMs. Furthermore, we observe that various alignment methods minimally resolve inconsistencies in the models' representation of disease prevalence across different languages. For further exploration and analysis, we make all data and a data visualization tool available at: www.crosscare.net.
Knowledge Graphs for the Life Sciences: Recent Developments, Challenges and Opportunities
Oct 06, 2023Abstract:The term life sciences refers to the disciplines that study living organisms and life processes, and include chemistry, biology, medicine, and a range of other related disciplines. Research efforts in life sciences are heavily data-driven, as they produce and consume vast amounts of scientific data, much of which is intrinsically relational and graph-structured. The volume of data and the complexity of scientific concepts and relations referred to therein promote the application of advanced knowledge-driven technologies for managing and interpreting data, with the ultimate aim to advance scientific discovery. In this survey and position paper, we discuss recent developments and advances in the use of graph-based technologies in life sciences and set out a vision for how these technologies will impact these fields into the future. We focus on three broad topics: the construction and management of Knowledge Graphs (KGs), the use of KGs and associated technologies in the discovery of new knowledge, and the use of KGs in artificial intelligence applications to support explanations (explainable AI). We select a few exemplary use cases for each topic, discuss the challenges and open research questions within these topics, and conclude with a perspective and outlook that summarizes the overarching challenges and their potential solutions as a guide for future research.
Ontology Pre-training for Poison Prediction
Jan 20, 2023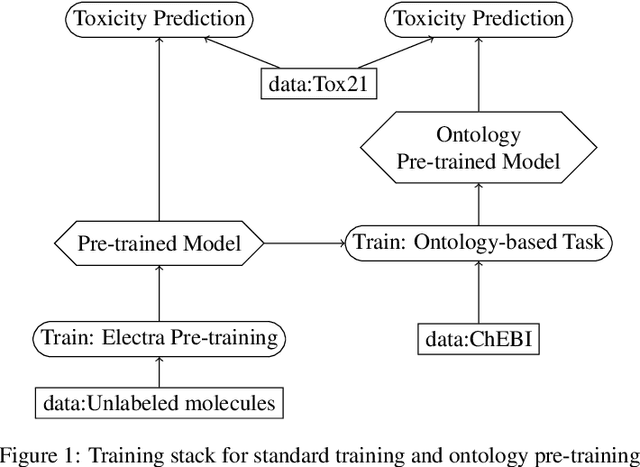
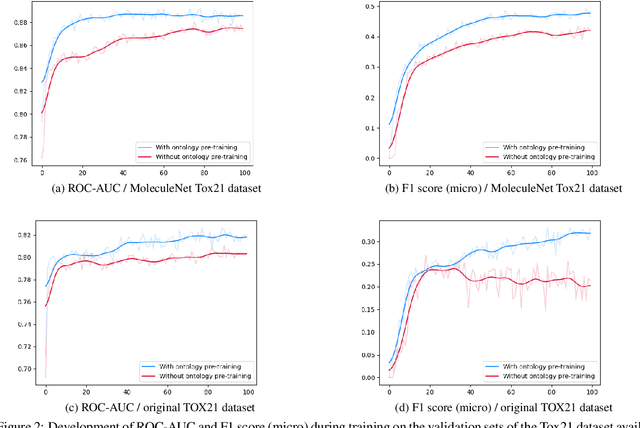
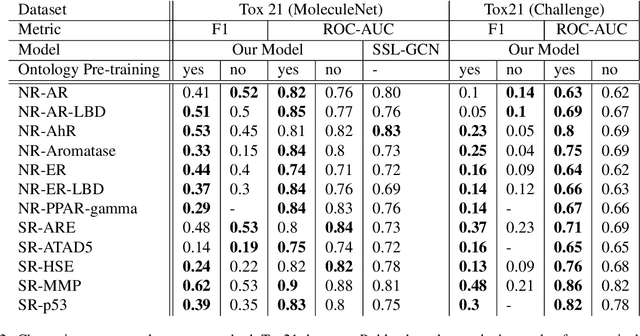
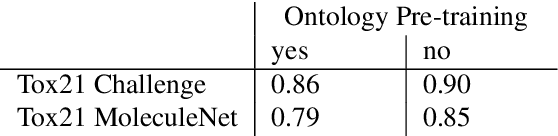
Abstract:Integrating human knowledge into neural networks has the potential to improve their robustness and interpretability. We have developed a novel approach to integrate knowledge from ontologies into the structure of a Transformer network which we call ontology pre-training: we train the network to predict membership in ontology classes as a way to embed the structure of the ontology into the network, and subsequently fine-tune the network for the particular prediction task. We apply this approach to a case study in predicting the potential toxicity of a small molecule based on its molecular structure, a challenging task for machine learning in life sciences chemistry. Our approach improves on the state of the art, and moreover has several additional benefits. First, we are able to show that the model learns to focus attention on more meaningful chemical groups when making predictions with ontology pre-training than without, paving a path towards greater robustness and interpretability. Second, the training time is reduced after ontology pre-training, indicating that the model is better placed to learn what matters for toxicity prediction with the ontology pre-training than without. This strategy has general applicability as a neuro-symbolic approach to embed meaningful semantics into neural networks.
When one Logic is Not Enough: Integrating First-order Annotations in OWL Ontologies
Oct 07, 2022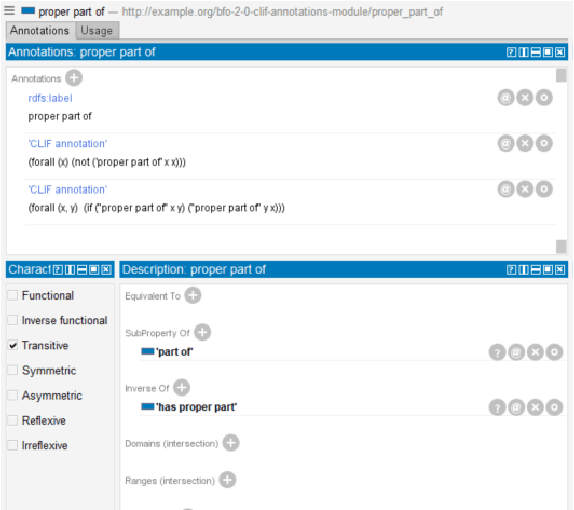
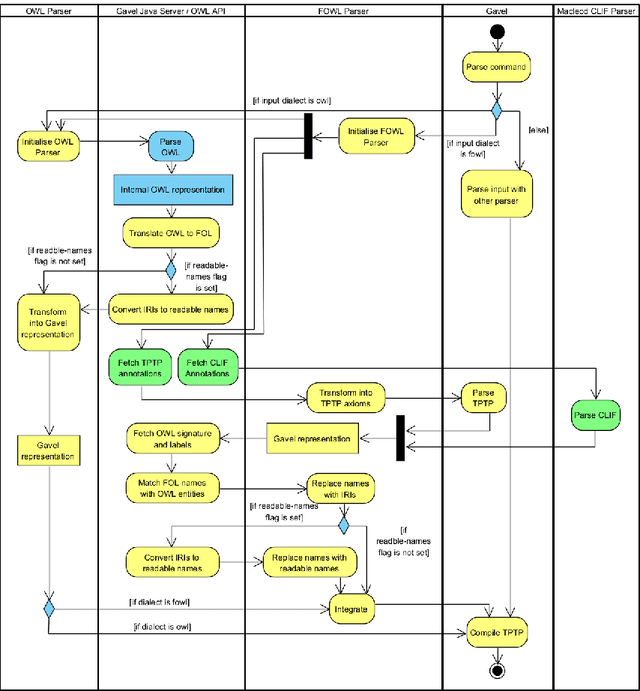

Abstract:In ontology development, there is a gap between domain ontologies which mostly use the web ontology language, OWL, and foundational ontologies written in first-order logic, FOL. To bridge this gap, we present Gavel, a tool that supports the development of heterogeneous 'FOWL' ontologies that extend OWL with FOL annotations, and is able to reason over the combined set of axioms. Since FOL annotations are stored in OWL annotations, FOWL ontologies remain compatible with the existing OWL infrastructure. We show that for the OWL domain ontology OBI, the stronger integration with its FOL top-level ontology BFO via our approach enables us to detect several inconsistencies. Furthermore, existing OWL ontologies can benefit from FOL annotations. We illustrate this with FOWL ontologies containing mereotopological axioms that enable new meaningful inferences. Finally, we show that even for large domain ontologies such as ChEBI, automatic reasoning with FOL annotations can be used to detect previously unnoticed errors in the classification.
ESC-Rules: Explainable, Semantically Constrained Rule Sets
Aug 26, 2022
Abstract:We describe a novel approach to explainable prediction of a continuous variable based on learning fuzzy weighted rules. Our model trains a set of weighted rules to maximise prediction accuracy and minimise an ontology-based 'semantic loss' function including user-specified constraints on the rules that should be learned in order to maximise the explainability of the resulting rule set from a user perspective. This system fuses quantitative sub-symbolic learning with symbolic learning and constraints based on domain knowledge. We illustrate our system on a case study in predicting the outcomes of behavioural interventions for smoking cessation, and show that it outperforms other interpretable approaches, achieving performance close to that of a deep learning model, while offering transparent explainability that is an essential requirement for decision-makers in the health domain.
Automated and Explainable Ontology Extension Based on Deep Learning: A Case Study in the Chemical Domain
Sep 19, 2021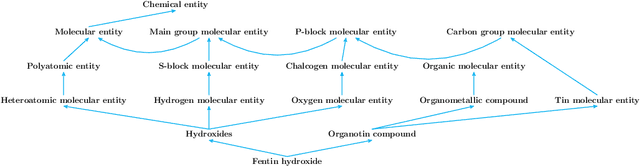
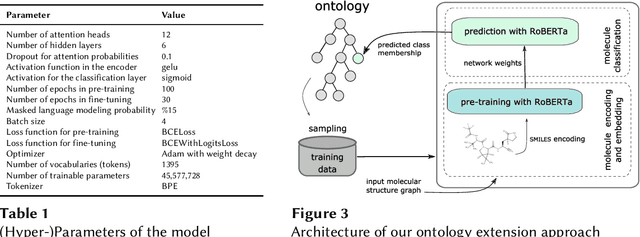
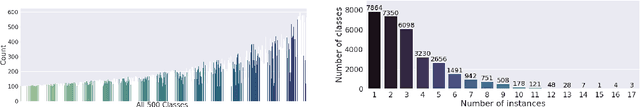
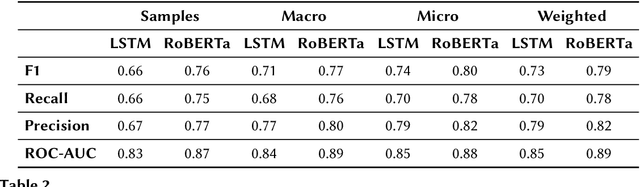
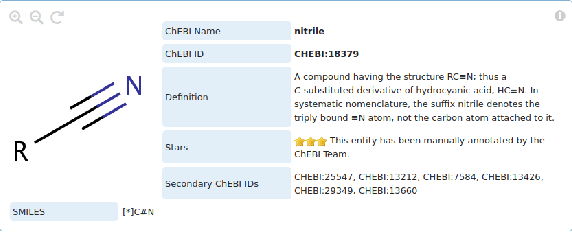
Abstract:Reference ontologies provide a shared vocabulary and knowledge resource for their domain. Manual construction enables them to maintain a high quality, allowing them to be widely accepted across their community. However, the manual development process does not scale for large domains. We present a new methodology for automatic ontology extension and apply it to the ChEBI ontology, a prominent reference ontology for life sciences chemistry. We trained a Transformer-based deep learning model on the leaf node structures from the ChEBI ontology and the classes to which they belong. The model is then capable of automatically classifying previously unseen chemical structures. The proposed model achieved an overall F1 score of 0.80, an improvement of 6 percentage points over our previous results on the same dataset. Additionally, we demonstrate how visualizing the model's attention weights can help to explain the results by providing insight into how the model made its decisions.
What's in an `is about' link? Chemical diagrams and the Information Artifact Ontology
Apr 21, 2012Abstract:The Information Artifact Ontology is an ontology in the domain of information entities. Core to the definition of what it is to be an information entity is the claim that an information entity must be `about' something, which is encoded in an axiom expressing that all information entities are about some entity. This axiom comes into conflict with ontological realism, since many information entities seem to be about non-existing entities, such as hypothetical molecules. We discuss this problem in the context of diagrams of molecules, a kind of information entity pervasively used throughout computational chemistry. We then propose a solution that recognizes that information entities such as diagrams are expressions of diagrammatic languages. In so doing, we not only address the problem of classifying diagrams that seem to be about non-existing entities but also allow a more sophisticated categorisation of information entities.
* 10 pages, 5 figures, presented at the 2nd International Conference on Biomedical Ontology (ICBO) 2011
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge