Martin Glauer
A semantic loss for ontology classification
May 03, 2024Abstract:Deep learning models are often unaware of the inherent constraints of the task they are applied to. However, many downstream tasks require logical consistency. For ontology classification tasks, such constraints include subsumption and disjointness relations between classes. In order to increase the consistency of deep learning models, we propose a semantic loss that combines label-based loss with terms penalising subsumption- or disjointness-violations. Our evaluation on the ChEBI ontology shows that the semantic loss is able to decrease the number of consistency violations by several orders of magnitude without decreasing the classification performance. In addition, we use the semantic loss for unsupervised learning. We show that this can further improve consistency on data from a distribution outside the scope of the supervised training.
Ontology Pre-training for Poison Prediction
Jan 20, 2023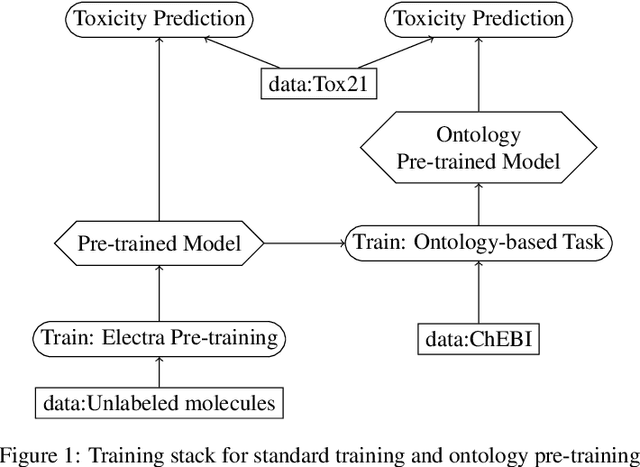
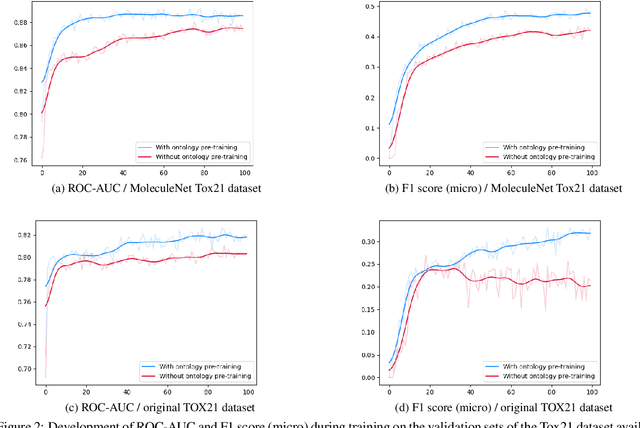
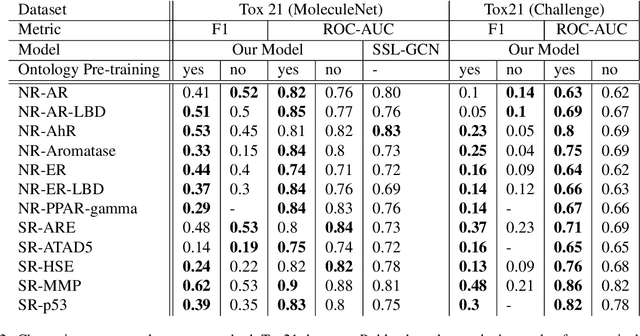
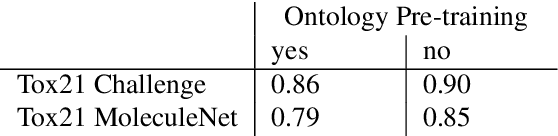
Abstract:Integrating human knowledge into neural networks has the potential to improve their robustness and interpretability. We have developed a novel approach to integrate knowledge from ontologies into the structure of a Transformer network which we call ontology pre-training: we train the network to predict membership in ontology classes as a way to embed the structure of the ontology into the network, and subsequently fine-tune the network for the particular prediction task. We apply this approach to a case study in predicting the potential toxicity of a small molecule based on its molecular structure, a challenging task for machine learning in life sciences chemistry. Our approach improves on the state of the art, and moreover has several additional benefits. First, we are able to show that the model learns to focus attention on more meaningful chemical groups when making predictions with ontology pre-training than without, paving a path towards greater robustness and interpretability. Second, the training time is reduced after ontology pre-training, indicating that the model is better placed to learn what matters for toxicity prediction with the ontology pre-training than without. This strategy has general applicability as a neuro-symbolic approach to embed meaningful semantics into neural networks.
When one Logic is Not Enough: Integrating First-order Annotations in OWL Ontologies
Oct 07, 2022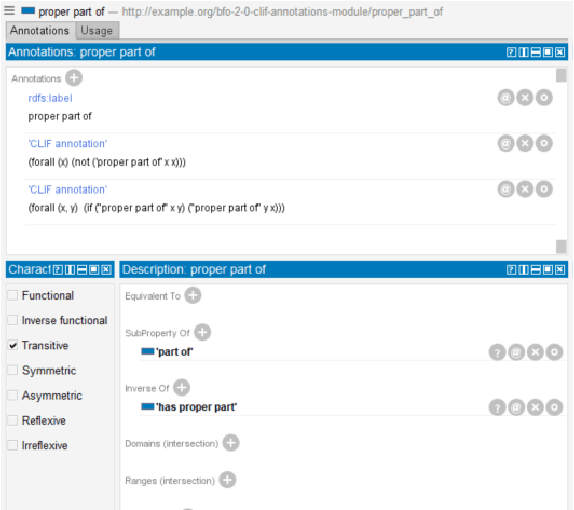
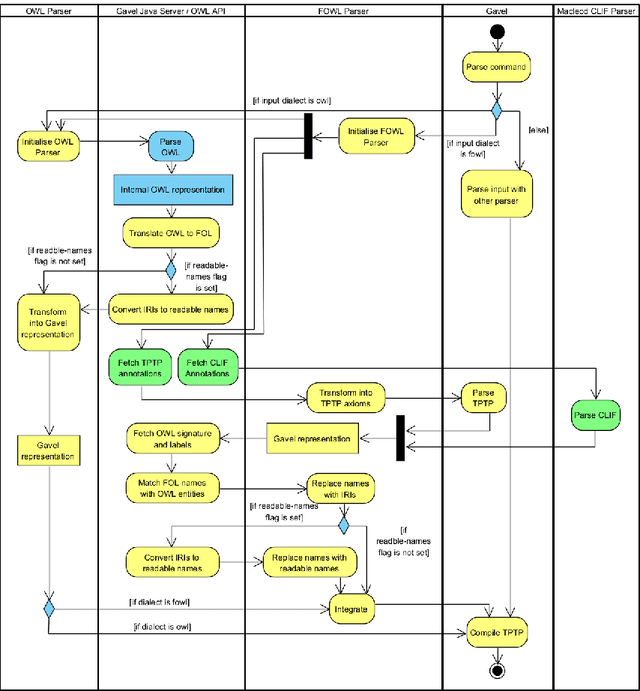

Abstract:In ontology development, there is a gap between domain ontologies which mostly use the web ontology language, OWL, and foundational ontologies written in first-order logic, FOL. To bridge this gap, we present Gavel, a tool that supports the development of heterogeneous 'FOWL' ontologies that extend OWL with FOL annotations, and is able to reason over the combined set of axioms. Since FOL annotations are stored in OWL annotations, FOWL ontologies remain compatible with the existing OWL infrastructure. We show that for the OWL domain ontology OBI, the stronger integration with its FOL top-level ontology BFO via our approach enables us to detect several inconsistencies. Furthermore, existing OWL ontologies can benefit from FOL annotations. We illustrate this with FOWL ontologies containing mereotopological axioms that enable new meaningful inferences. Finally, we show that even for large domain ontologies such as ChEBI, automatic reasoning with FOL annotations can be used to detect previously unnoticed errors in the classification.
ESC-Rules: Explainable, Semantically Constrained Rule Sets
Aug 26, 2022
Abstract:We describe a novel approach to explainable prediction of a continuous variable based on learning fuzzy weighted rules. Our model trains a set of weighted rules to maximise prediction accuracy and minimise an ontology-based 'semantic loss' function including user-specified constraints on the rules that should be learned in order to maximise the explainability of the resulting rule set from a user perspective. This system fuses quantitative sub-symbolic learning with symbolic learning and constraints based on domain knowledge. We illustrate our system on a case study in predicting the outcomes of behavioural interventions for smoking cessation, and show that it outperforms other interpretable approaches, achieving performance close to that of a deep learning model, while offering transparent explainability that is an essential requirement for decision-makers in the health domain.
Automated and Explainable Ontology Extension Based on Deep Learning: A Case Study in the Chemical Domain
Sep 19, 2021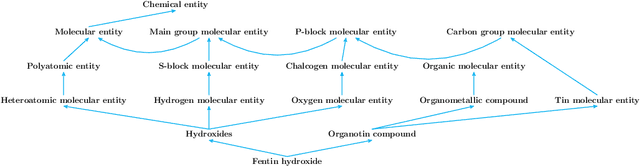
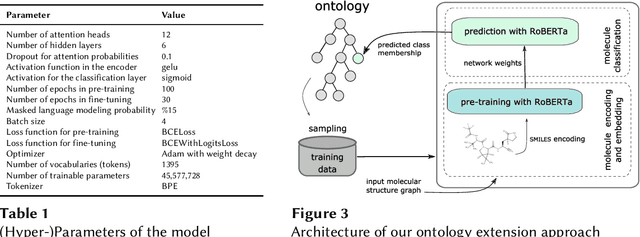
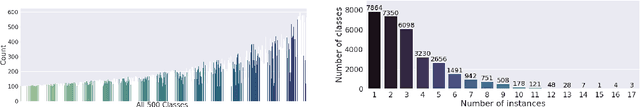
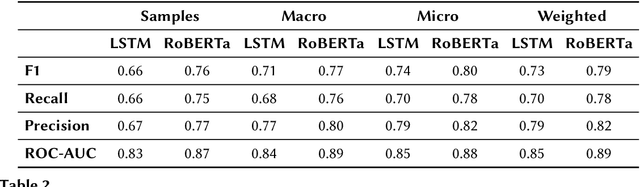
Abstract:Reference ontologies provide a shared vocabulary and knowledge resource for their domain. Manual construction enables them to maintain a high quality, allowing them to be widely accepted across their community. However, the manual development process does not scale for large domains. We present a new methodology for automatic ontology extension and apply it to the ChEBI ontology, a prominent reference ontology for life sciences chemistry. We trained a Transformer-based deep learning model on the leaf node structures from the ChEBI ontology and the classes to which they belong. The model is then capable of automatically classifying previously unseen chemical structures. The proposed model achieved an overall F1 score of 0.80, an improvement of 6 percentage points over our previous results on the same dataset. Additionally, we demonstrate how visualizing the model's attention weights can help to explain the results by providing insight into how the model made its decisions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge