Ivan Lerner
Spectral structure learning for clinical time series
Feb 17, 2025
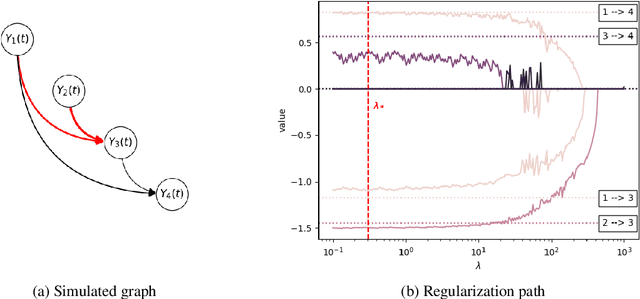

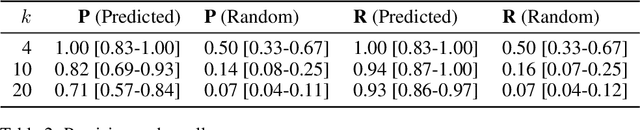
Abstract:We develop and evaluate a structure learning algorithm for clinical time series. Clinical time series are multivariate time series observed in multiple patients and irregularly sampled, challenging existing structure learning algorithms. We assume that our times series are realizations of StructGP, a k-dimensional multi-output or multi-task stationary Gaussian process (GP), with independent patients sharing the same covariance function. StructGP encodes ordered conditional relations between time series, represented in a directed acyclic graph. We implement an adapted NOTEARS algorithm, which based on a differentiable definition of acyclicity, recovers the graph by solving a series of continuous optimization problems. Simulation results show that up to mean degree 3 and 20 tasks, we reach a median recall of 0.93% [IQR, 0.86, 0.97] while keeping a median precision of 0.71% [0.57-0.84], for recovering directed edges. We further show that the regularization path is key to identifying the graph. With StructGP, we proposed a model of time series dependencies, that flexibly adapt to different time series regularity, while enabling us to learn these dependencies from observations.
Facilitating phenotyping from clinical texts: the medkit library
Aug 30, 2024

Abstract:Phenotyping consists in applying algorithms to identify individuals associated with a specific, potentially complex, trait or condition, typically out of a collection of Electronic Health Records (EHRs). Because a lot of the clinical information of EHRs are lying in texts, phenotyping from text takes an important role in studies that rely on the secondary use of EHRs. However, the heterogeneity and highly specialized aspect of both the content and form of clinical texts makes this task particularly tedious, and is the source of time and cost constraints in observational studies. To facilitate the development, evaluation and reproductibility of phenotyping pipelines, we developed an open-source Python library named medkit. It enables composing data processing pipelines made of easy-to-reuse software bricks, named medkit operations. In addition to the core of the library, we share the operations and pipelines we already developed and invite the phenotyping community for their reuse and enrichment. medkit is available at https://github.com/medkit-lib/medkit
Learning the grammar of prescription: recurrent neural network grammars for medication information extraction in clinical texts
Apr 24, 2020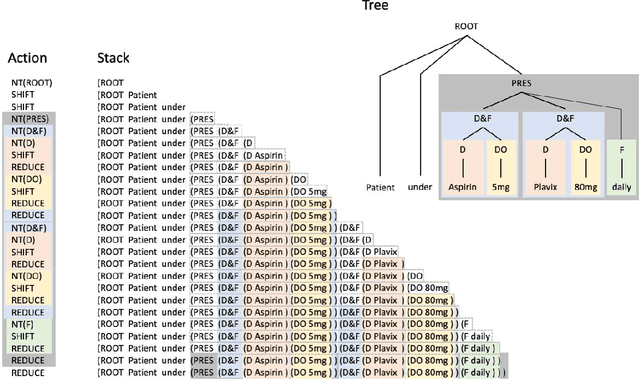
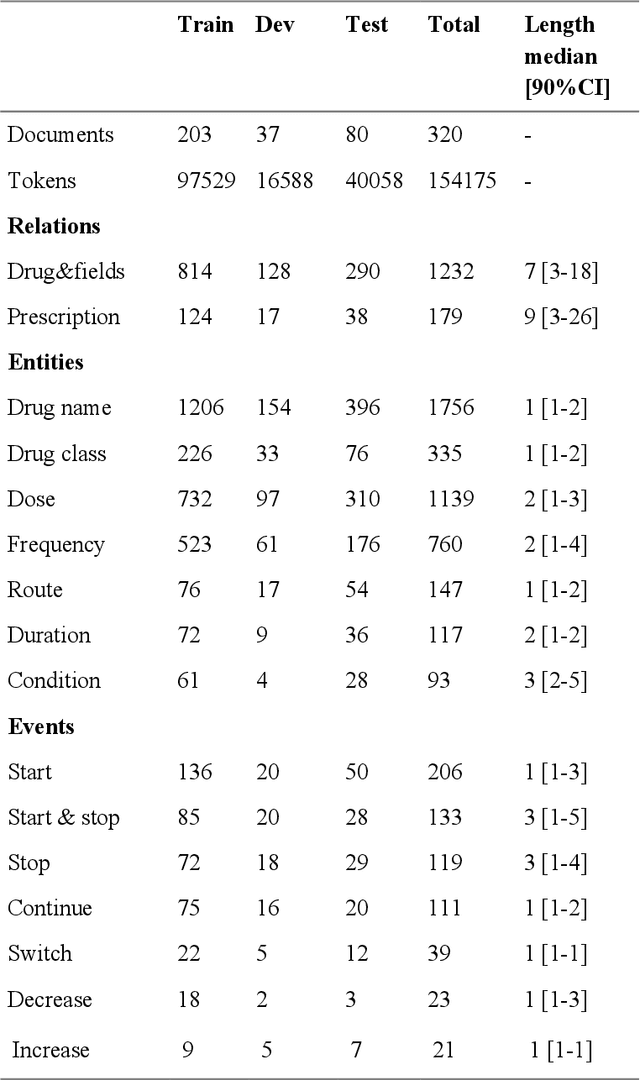
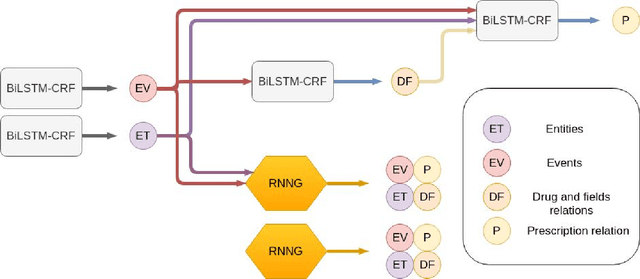
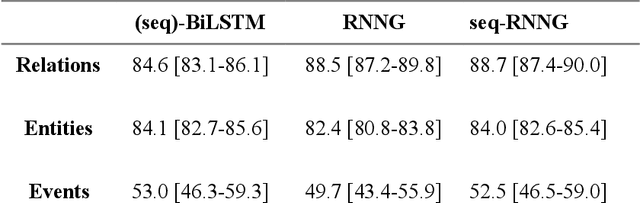
Abstract:In this study, we evaluated the RNNG, a neural top-down transition based parser, for medication information extraction in clinical texts. We evaluated this model on a French clinical corpus. The task was to extract the name of a drug (or class of drug), as well as fields informing its administration: frequency, dosage, duration, condition and route of administration. We compared the RNNG model that jointly identify entities and their relations with separate BiLSTMs models for entities and relations as baselines. We call seq-BiLSTMs the baseline models for relations extraction that takes as extra-input the output of the BiLSTMs for entities. RNNG outperforms seq-BiLSTM for identifying relations, with on average 88.5% [87.2-89.8] versus 84.6 [83.1-86.1] F-measure. However, RNNG is weaker than the baseline BiLSTM on detecting entities, with on average 82.4 [80.8-83.8] versus 84.1 [82.7-85.6] % F- measure. RNNG trained only for detecting relations is weaker than RNNG with the joint modelling objective, 87.4 [85.8-88.8] versus 88.5% [87.2-89.8]. The performance of RNNG on relations can be explained both by the model architecture, which provides shortcut between distant parts of the sentence, and the joint modelling objective which allow the RNNG to learn richer representations. RNNG is efficient for modeling relations between entities in medical texts and its performances are close to those of a BiLSTM for entity detection.
Terminologies augmented recurrent neural network model for clinical named entity recognition
May 15, 2019



Abstract:We aimed to enhance the performance of a supervised model for clinical named-entity recognition (NER) using medical terminologies. In order to evaluate our system in French, we built a corpus for 5 types of clinical entities. We used a terminology-based system as baseline, built upon UMLS and SNOMED. Then, we evaluated a biGRU-CRF, and an hybrid system using the prediction of the terminology-based system as feature for the biGRU-CRF. In English, we evaluated the NER systems on the i2b2-2009 Medication Challenge for Drug name recognition, which contained 8,573 entities for 268 documents. In French, we built APcNER, a corpus of 147 documents annotated for 5 entities (drug name, sign or symptom, disease or disorder, diagnostic procedure or lab test and therapeutic procedure). We evaluated each NER systems using exact and partial match definition of F-measure for NER. The APcNER contains 4,837 entities which took 28 hours to annotate, the inter-annotator agreement was acceptable for Drug name in exact match (85%) and acceptable for other entity types in non-exact match (>70%). For drug name recognition on both i2b2-2009 and APcNER, the biGRU-CRF performed better that the terminology-based system, with an exact-match F-measure of 91.1% versus 73% and 81.9% versus 75% respectively. Moreover, the hybrid system outperformed the biGRU-CRF, with an exact-match F-measure of 92.2% versus 91.1% (i2b2-2009) and 88.4% versus 81.9% (APcNER). On APcNER corpus, the micro-average F-measure of the hybrid system on the 5 entities was 69.5% in exact match, and 84.1% in non-exact match. APcNER is a French corpus for clinical-NER of five type of entities which covers a large variety of document types. Extending supervised model with terminology allowed for an easy performance gain, especially in low regimes of entities, and established near state of the art results on the i2b2-2009 corpus.
Hybrid Approaches for our Participation to the n2c2 Challenge on Cohort Selection for Clinical Trials
Mar 19, 2019

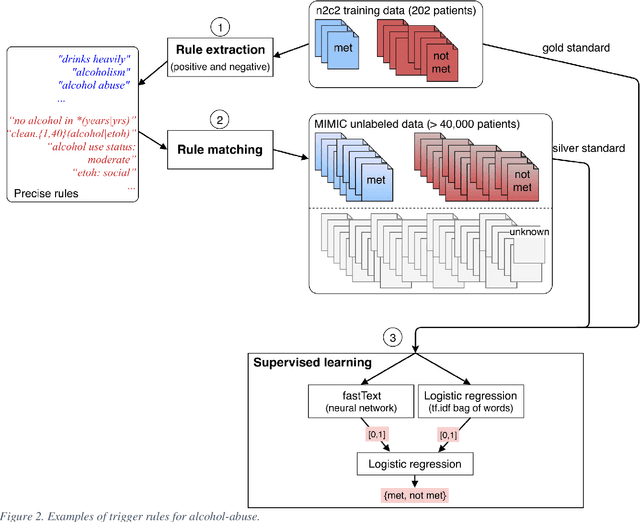
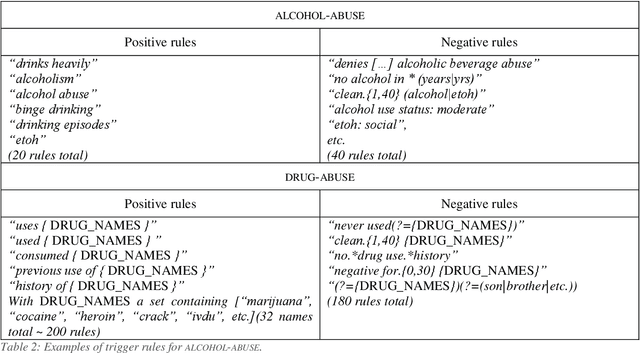
Abstract:Objective: Natural language processing can help minimize human intervention in identifying patients meeting eligibility criteria for clinical trials, but there is still a long way to go to obtain a general and systematic approach that is useful for researchers. We describe two methods taking a step in this direction and present their results obtained during the n2c2 challenge on cohort selection for clinical trials. Materials and Methods: The first method is a weakly supervised method using an unlabeled corpus (MIMIC) to build a silver standard, by producing semi-automatically a small and very precise set of rules to detect some samples of positive and negative patients. This silver standard is then used to train a traditional supervised model. The second method is a terminology-based approach where a medical expert selects the appropriate concepts, and a procedure is defined to search the terms and check the structural or temporal constraints. Results: On the n2c2 dataset containing annotated data about 13 selection criteria on 288 patients, we obtained an overall F1-measure of 0.8969, which is the third best result out of 45 participant teams, with no statistically significant difference with the best-ranked team. Discussion: Both approaches obtained very encouraging results and apply to different types of criteria. The weakly supervised method requires explicit descriptions of positive and negative examples in some reports. The terminology-based method is very efficient when medical concepts carry most of the relevant information. Conclusion: It is unlikely that much more annotated data will be soon available for the task of identifying a wide range of patient phenotypes. One must focus on weakly or non-supervised learning methods using both structured and unstructured data and relying on a comprehensive representation of the patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge