Learning the grammar of prescription: recurrent neural network grammars for medication information extraction in clinical texts
Paper and Code
Apr 24, 2020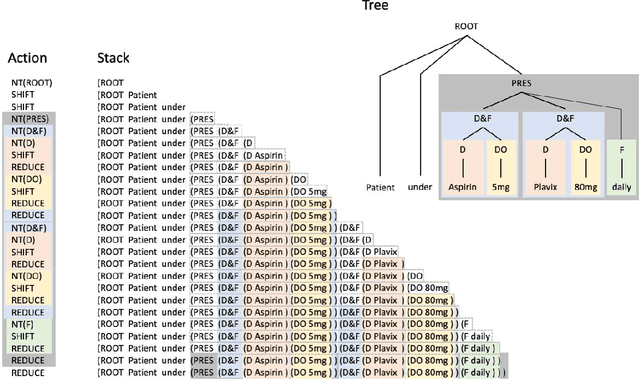
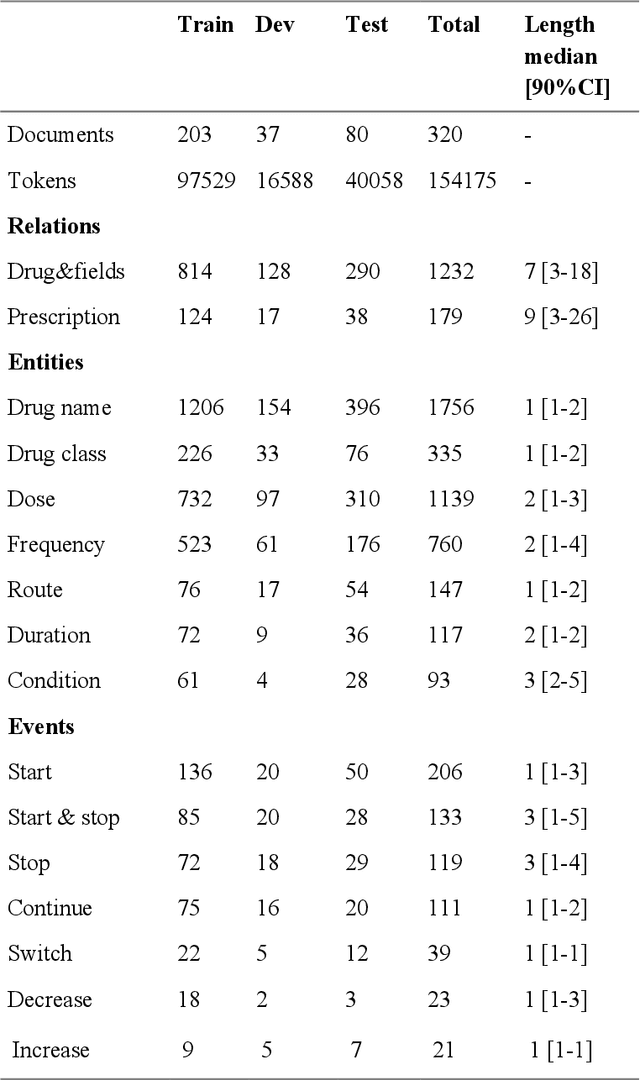
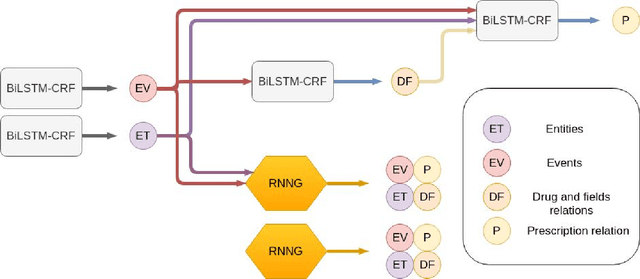
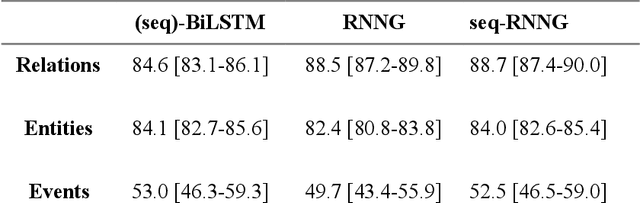
In this study, we evaluated the RNNG, a neural top-down transition based parser, for medication information extraction in clinical texts. We evaluated this model on a French clinical corpus. The task was to extract the name of a drug (or class of drug), as well as fields informing its administration: frequency, dosage, duration, condition and route of administration. We compared the RNNG model that jointly identify entities and their relations with separate BiLSTMs models for entities and relations as baselines. We call seq-BiLSTMs the baseline models for relations extraction that takes as extra-input the output of the BiLSTMs for entities. RNNG outperforms seq-BiLSTM for identifying relations, with on average 88.5% [87.2-89.8] versus 84.6 [83.1-86.1] F-measure. However, RNNG is weaker than the baseline BiLSTM on detecting entities, with on average 82.4 [80.8-83.8] versus 84.1 [82.7-85.6] % F- measure. RNNG trained only for detecting relations is weaker than RNNG with the joint modelling objective, 87.4 [85.8-88.8] versus 88.5% [87.2-89.8]. The performance of RNNG on relations can be explained both by the model architecture, which provides shortcut between distant parts of the sentence, and the joint modelling objective which allow the RNNG to learn richer representations. RNNG is efficient for modeling relations between entities in medical texts and its performances are close to those of a BiLSTM for entity detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge