Guy Fagherazzi
Identifying causal associations in tweets using deep learning: Use case on diabetes-related tweets from 2017-2021
Nov 04, 2021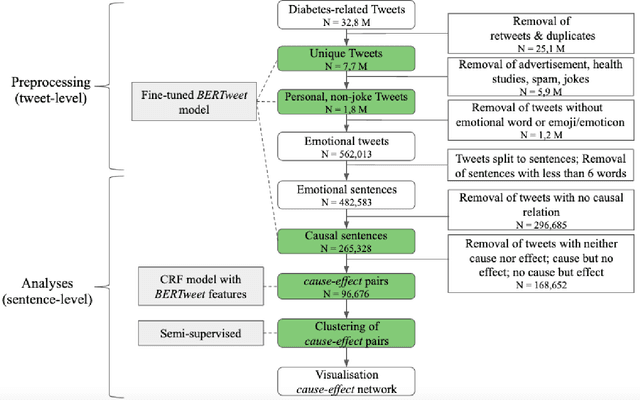
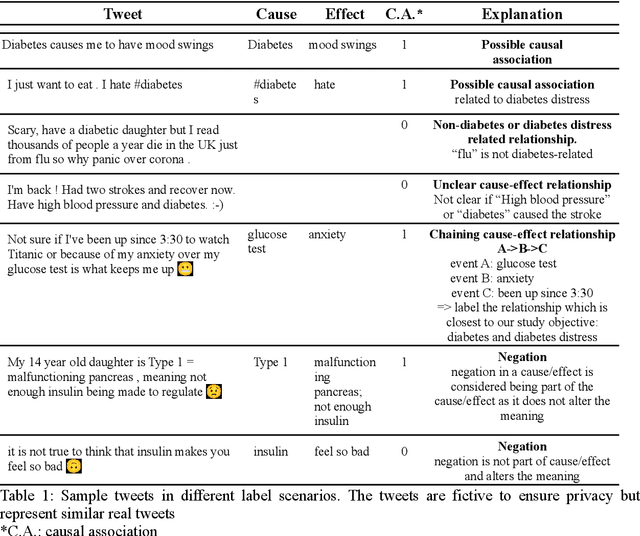
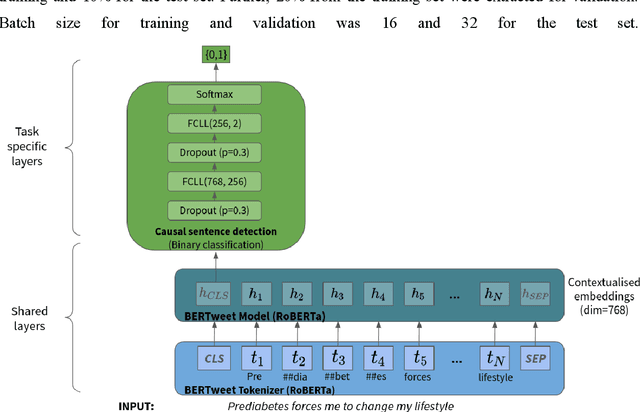
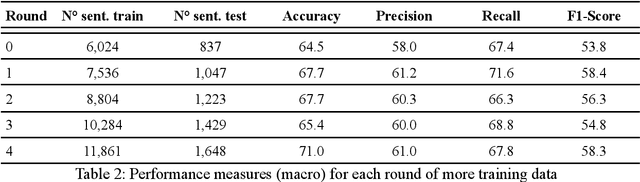
Abstract:Objective: Leveraging machine learning methods, we aim to extract both explicit and implicit cause-effect associations in patient-reported, diabetes-related tweets and provide a tool to better understand opinion, feelings and observations shared within the diabetes online community from a causality perspective. Materials and Methods: More than 30 million diabetes-related tweets in English were collected between April 2017 and January 2021. Deep learning and natural language processing methods were applied to focus on tweets with personal and emotional content. A cause-effect-tweet dataset was manually labeled and used to train 1) a fine-tuned Bertweet model to detect causal sentences containing a causal association 2) a CRF model with BERT based features to extract possible cause-effect associations. Causes and effects were clustered in a semi-supervised approach and visualised in an interactive cause-effect-network. Results: Causal sentences were detected with a recall of 68% in an imbalanced dataset. A CRF model with BERT based features outperformed a fine-tuned BERT model for cause-effect detection with a macro recall of 68%. This led to 96,676 sentences with cause-effect associations. "Diabetes" was identified as the central cluster followed by "Death" and "Insulin". Insulin pricing related causes were frequently associated with "Death". Conclusions: A novel methodology was developed to detect causal sentences and identify both explicit and implicit, single and multi-word cause and corresponding effect as expressed in diabetes-related tweets leveraging BERT-based architectures and visualised as cause-effect-network. Extracting causal associations on real-life, patient reported outcomes in social media data provides a useful complementary source of information in diabetes research.
Deep clustering of longitudinal data
Feb 09, 2018



Abstract:Deep neural networks are a family of computational models that have led to a dramatical improvement of the state of the art in several domains such as image, voice or text analysis. These methods provide a framework to model complex, non-linear interactions in large datasets, and are naturally suited to the analysis of hierarchical data such as, for instance, longitudinal data with the use of recurrent neural networks. In the other hand, cohort studies have become a tool of importance in the research field of epidemiology. In such studies, variables are measured repeatedly over time, to allow the practitioner to study their temporal evolution as trajectories, and, as such, as longitudinal data. This paper investigates the application of the advanced modelling techniques provided by the deep learning framework in the analysis of the longitudinal data provided by cohort studies. Methods: A method for visualizing and clustering longitudinal dataset is proposed, and compared to other widely used approaches to the problem on both real and simulated datasets. Results: The proposed method is shown to be coherent with the preexisting procedures on simple tasks, and to outperform them on more complex tasks such as the partitioning of longitudinal datasets into non-spherical clusters. Conclusion: Deep artificial neural networks can be used to visualize longitudinal data in a low dimensional manifold that is much simpler to interpret than traditional longitudinal plots are. Consequently, practitioners should start considering the use of deep artificial neural networks for the analysis of their longitudinal data in studies to come.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge