Domenec Puig
UltrasODM: A Dual Stream Optical Flow Mamba Network for 3D Freehand Ultrasound Reconstruction
Dec 08, 2025Abstract:Clinical ultrasound acquisition is highly operator-dependent, where rapid probe motion and brightness fluctuations often lead to reconstruction errors that reduce trust and clinical utility. We present UltrasODM, a dual-stream framework that assists sonographers during acquisition through calibrated per-frame uncertainty, saliency-based diagnostics, and actionable prompts. UltrasODM integrates (i) a contrastive ranking module that groups frames by motion similarity, (ii) an optical-flow stream fused with Dual-Mamba temporal modules for robust 6-DoF pose estimation, and (iii) a Human-in-the-Loop (HITL) layer combining Bayesian uncertainty, clinician-calibrated thresholds, and saliency maps highlighting regions of low confidence. When uncertainty exceeds the threshold, the system issues unobtrusive alerts suggesting corrective actions such as re-scanning highlighted regions or slowing the sweep. Evaluated on a clinical freehand ultrasound dataset, UltrasODM reduces drift by 15.2%, distance error by 12.1%, and Hausdorff distance by 10.1% relative to UltrasOM, while producing per-frame uncertainty and saliency outputs. By emphasizing transparency and clinician feedback, UltrasODM improves reconstruction reliability and supports safer, more trustworthy clinical workflows. Our code is publicly available at https://github.com/AnandMayank/UltrasODM.
M-TabNet: A Multi-Encoder Transformer Model for Predicting Neonatal Birth Weight from Multimodal Data
Apr 20, 2025Abstract:Birth weight (BW) is a key indicator of neonatal health, with low birth weight (LBW) linked to increased mortality and morbidity. Early prediction of BW enables timely interventions; however, current methods like ultrasonography have limitations, including reduced accuracy before 20 weeks and operator dependent variability. Existing models often neglect nutritional and genetic influences, focusing mainly on physiological and lifestyle factors. This study presents an attention-based transformer model with a multi-encoder architecture for early (less than 12 weeks of gestation) BW prediction. Our model effectively integrates diverse maternal data such as physiological, lifestyle, nutritional, and genetic, addressing limitations seen in prior attention-based models such as TabNet. The model achieves a Mean Absolute Error (MAE) of 122 grams and an R-squared value of 0.94, demonstrating high predictive accuracy and interoperability with our in-house private dataset. Independent validation confirms generalizability (MAE: 105 grams, R-squared: 0.95) with the IEEE children dataset. To enhance clinical utility, predicted BW is classified into low and normal categories, achieving a sensitivity of 97.55% and a specificity of 94.48%, facilitating early risk stratification. Model interpretability is reinforced through feature importance and SHAP analyses, highlighting significant influences of maternal age, tobacco exposure, and vitamin B12 status, with genetic factors playing a secondary role. Our results emphasize the potential of advanced deep-learning models to improve early BW prediction, offering clinicians a robust, interpretable, and personalized tool for identifying pregnancies at risk and optimizing neonatal outcomes.
FGR-Net:Interpretable fundus imagegradeability classification based on deepreconstruction learning
Sep 16, 2024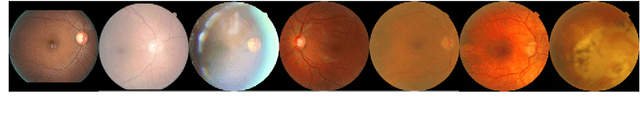
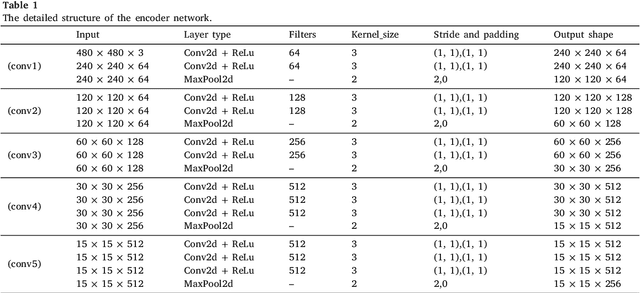
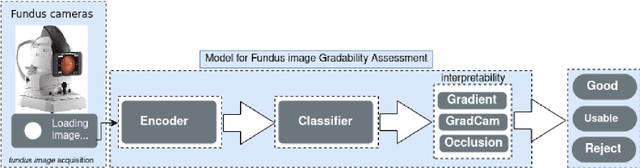
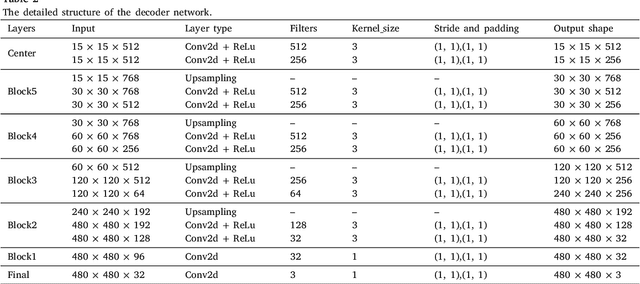
Abstract:The performance of diagnostic Computer-Aided Design (CAD) systems for retinal diseases depends on the quality of the retinal images being screened. Thus, many studies have been developed to evaluate and assess the quality of such retinal images. However, most of them did not investigate the relationship between the accuracy of the developed models and the quality of the visualization of interpretability methods for distinguishing between gradable and non-gradable retinal images. Consequently, this paper presents a novel framework called FGR-Net to automatically assess and interpret underlying fundus image quality by merging an autoencoder network with a classifier network. The FGR-Net model also provides an interpretable quality assessment through visualizations. In particular, FGR-Net uses a deep autoencoder to reconstruct the input image in order to extract the visual characteristics of the input fundus images based on self-supervised learning. The extracted features by the autoencoder are then fed into a deep classifier network to distinguish between gradable and ungradable fundus images. FGR-Net is evaluated with different interpretability methods, which indicates that the autoencoder is a key factor in forcing the classifier to focus on the relevant structures of the fundus images, such as the fovea, optic disk, and prominent blood vessels. Additionally, the interpretability methods can provide visual feedback for ophthalmologists to understand how our model evaluates the quality of fundus images. The experimental results showed the superiority of FGR-Net over the state-of-the-art quality assessment methods, with an accuracy of 89% and an F1-score of 87%.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024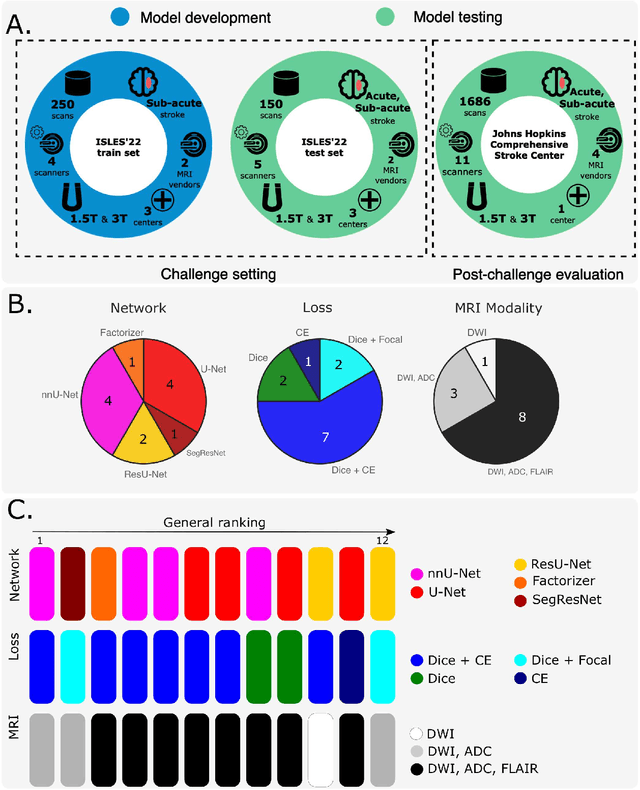
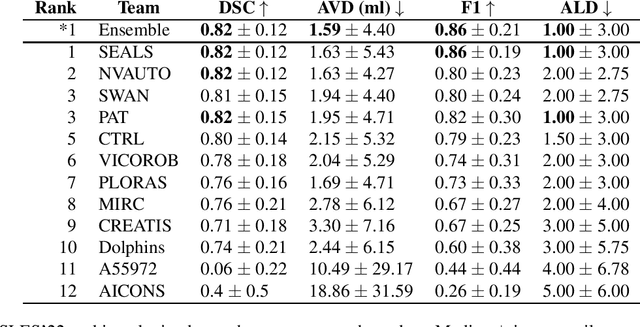
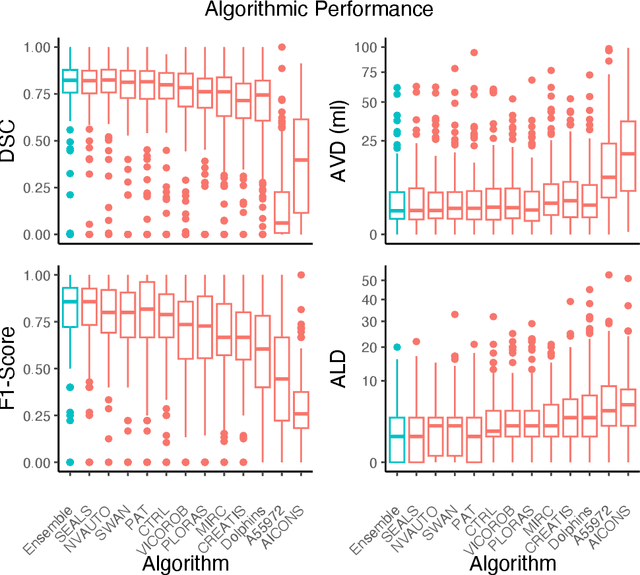
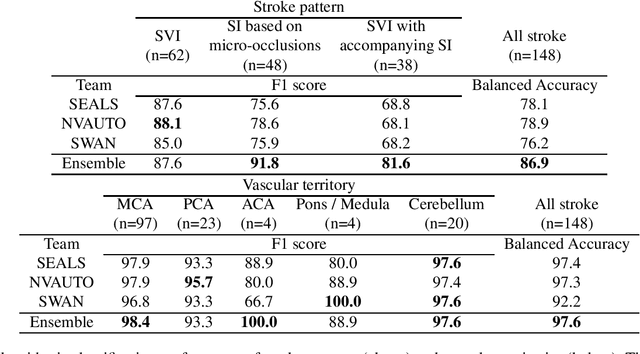
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Knowledge Distillation for Adaptive MRI Prostate Segmentation Based on Limit-Trained Multi-Teacher Models
Mar 16, 2023Abstract:With numerous medical tasks, the performance of deep models has recently experienced considerable improvements. These models are often adept learners. Yet, their intricate architectural design and high computational complexity make deploying them in clinical settings challenging, particularly with devices with limited resources. To deal with this issue, Knowledge Distillation (KD) has been proposed as a compression method and an acceleration technology. KD is an efficient learning strategy that can transfer knowledge from a burdensome model (i.e., teacher model) to a lightweight model (i.e., student model). Hence we can obtain a compact model with low parameters with preserving the teacher's performance. Therefore, we develop a KD-based deep model for prostate MRI segmentation in this work by combining features-based distillation with Kullback-Leibler divergence, Lovasz, and Dice losses. We further demonstrate its effectiveness by applying two compression procedures: 1) distilling knowledge to a student model from a single well-trained teacher, and 2) since most of the medical applications have a small dataset, we train multiple teachers that each one trained with a small set of images to learn an adaptive student model as close to the teachers as possible considering the desired accuracy and fast inference time. Extensive experiments were conducted on a public multi-site prostate tumor dataset, showing that the proposed adaptation KD strategy improves the dice similarity score by 9%, outperforming all tested well-established baseline models.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022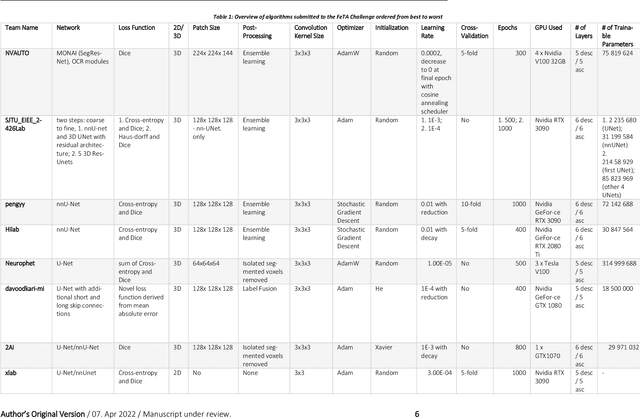
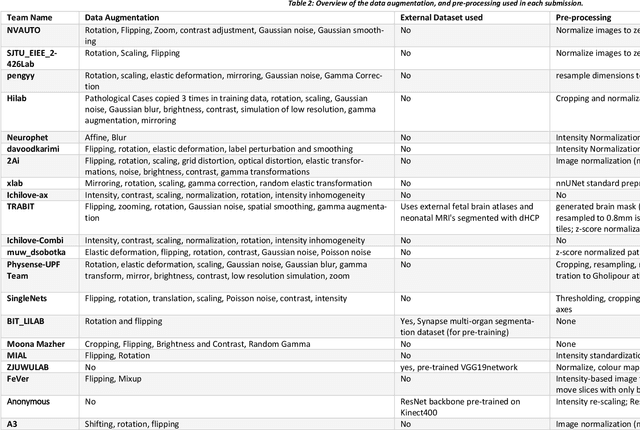
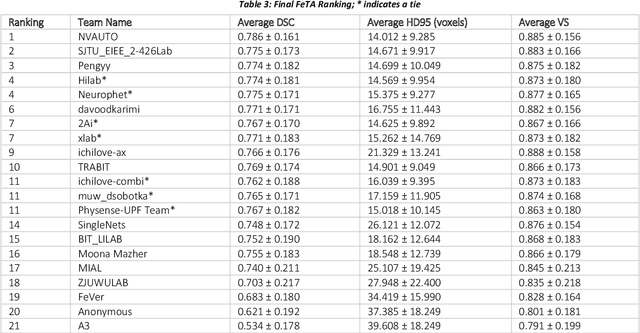
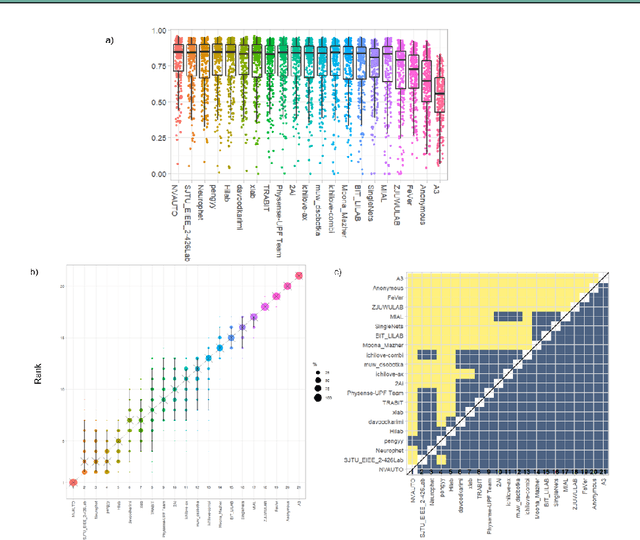
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
GCNDepth: Self-supervised Monocular Depth Estimation based on Graph Convolutional Network
Dec 13, 2021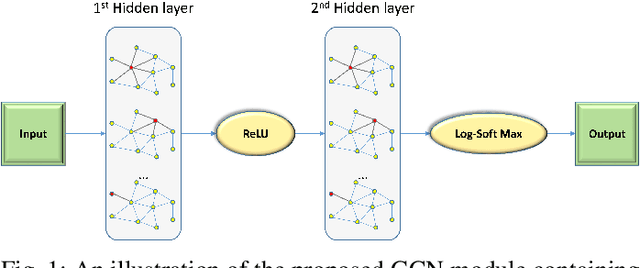
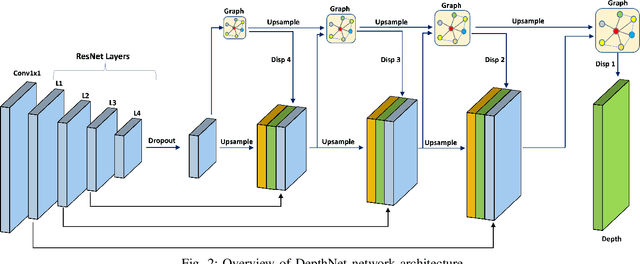
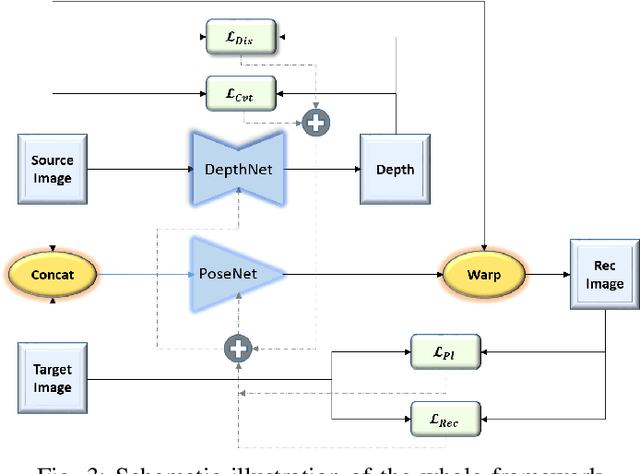
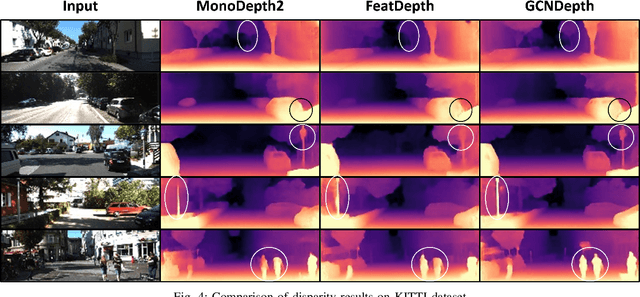
Abstract:Depth estimation is a challenging task of 3D reconstruction to enhance the accuracy sensing of environment awareness. This work brings a new solution with a set of improvements, which increase the quantitative and qualitative understanding of depth maps compared to existing methods. Recently, a convolutional neural network (CNN) has demonstrated its extraordinary ability in estimating depth maps from monocular videos. However, traditional CNN does not support topological structure and they can work only on regular image regions with determined size and weights. On the other hand, graph convolutional networks (GCN) can handle the convolution on non-Euclidean data and it can be applied to irregular image regions within a topological structure. Therefore, in this work in order to preserve object geometric appearances and distributions, we aim at exploiting GCN for a self-supervised depth estimation model. Our model consists of two parallel auto-encoder networks: the first is an auto-encoder that will depend on ResNet-50 and extract the feature from the input image and on multi-scale GCN to estimate the depth map. In turn, the second network will be used to estimate the ego-motion vector (i.e., 3D pose) between two consecutive frames based on ResNet-18. Both the estimated 3D pose and depth map will be used for constructing a target image. A combination of loss functions related to photometric, projection, and smoothness is used to cope with bad depth prediction and preserve the discontinuities of the objects. In particular, our method provided comparable and promising results with a high prediction accuracy of 89% on the publicly KITTI and Make3D datasets along with a reduction of 40% in the number of trainable parameters compared to the state of the art solutions. The source code is publicly available at https://github.com/ArminMasoumian/GCNDepth.git
Absolute distance prediction based on deep learning object detection and monocular depth estimation models
Nov 02, 2021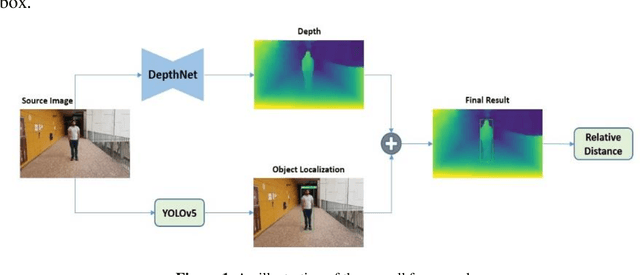
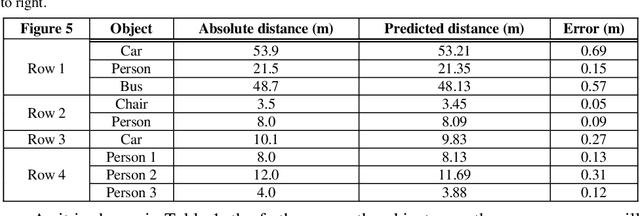
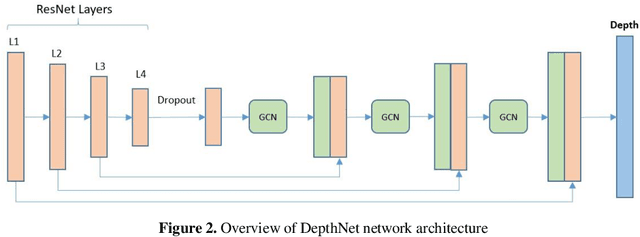
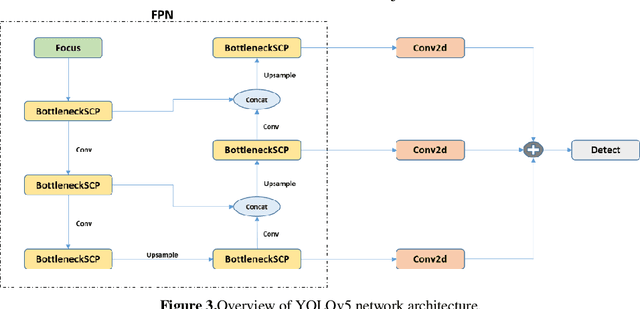
Abstract:Determining the distance between the objects in a scene and the camera sensor from 2D images is feasible by estimating depth images using stereo cameras or 3D cameras. The outcome of depth estimation is relative distances that can be used to calculate absolute distances to be applicable in reality. However, distance estimation is very challenging using 2D monocular cameras. This paper presents a deep learning framework that consists of two deep networks for depth estimation and object detection using a single image. Firstly, objects in the scene are detected and localized using the You Only Look Once (YOLOv5) network. In parallel, the estimated depth image is computed using a deep autoencoder network to detect the relative distances. The proposed object detection based YOLO was trained using a supervised learning technique, in turn, the network of depth estimation was self-supervised training. The presented distance estimation framework was evaluated on real images of outdoor scenes. The achieved results show that the proposed framework is promising and it yields an accuracy of 96% with RMSE of 0.203 of the correct absolute distance.
AWEU-Net: An Attention-Aware Weight Excitation U-Net for Lung Nodule Segmentation
Oct 11, 2021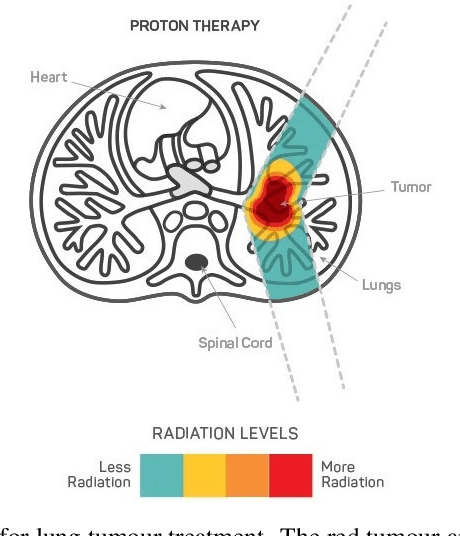
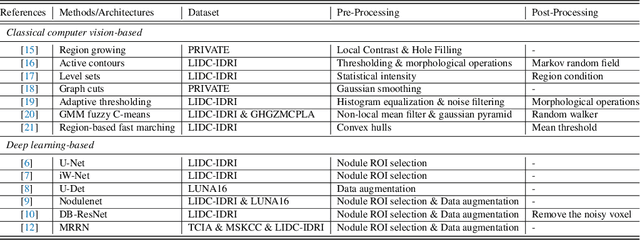
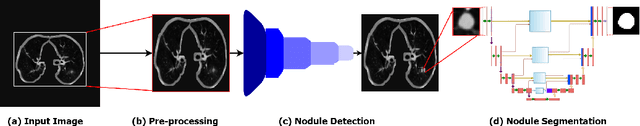
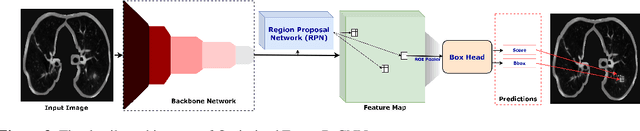
Abstract:Lung cancer is deadly cancer that causes millions of deaths every year around the world. Accurate lung nodule detection and segmentation in computed tomography (CT) images is the most important part of diagnosing lung cancer in the early stage. Most of the existing systems are semi-automated and need to manually select the lung and nodules regions to perform the segmentation task. To address these challenges, we proposed a fully automated end-to-end lung nodule detection and segmentation system based on a deep learning approach. In this paper, we used Optimized Faster R-CNN; a state-of-the-art detection model to detect the lung nodule regions in the CT scans. Furthermore, we proposed an attention-aware weight excitation U-Net, called AWEU-Net, for lung nodule segmentation and boundaries detection. To achieve more accurate nodule segmentation, in AWEU-Net, we proposed position attention-aware weight excitation (PAWE), and channel attention-aware weight excitation (CAWE) blocks to highlight the best aligned spatial and channel features in the input feature maps. The experimental results demonstrate that our proposed model yields a Dice score of 89.79% and 90.35%, and an intersection over union (IoU) of 82.34% and 83.21% on the publicly LUNA16 and LIDC-IDRI datasets, respectively.
Adversarial Learning with Multiscale Features and Kernel Factorization for Retinal Blood Vessel Segmentation
Jul 05, 2019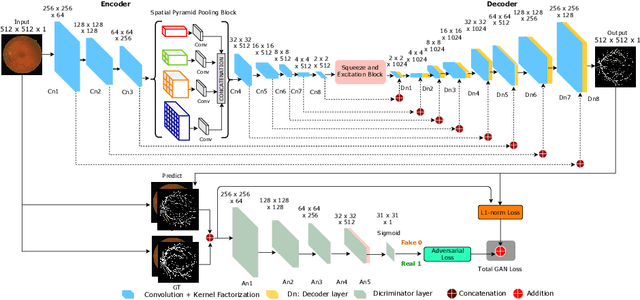
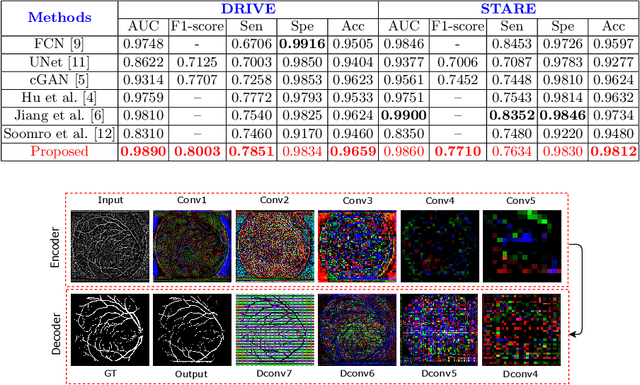
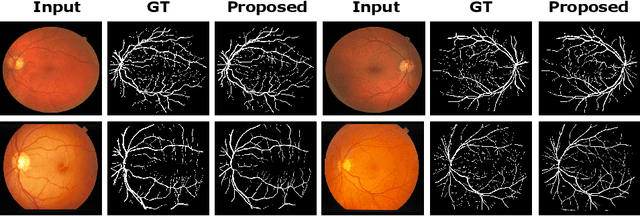
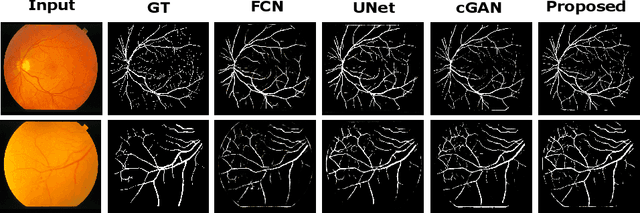
Abstract:In this paper, we propose an efficient blood vessel segmentation method for the eye fundus images using adversarial learning with multiscale features and kernel factorization. In the generator network of the adversarial framework, spatial pyramid pooling, kernel factorization and squeeze excitation block are employed to enhance the feature representation in spatial domain on different scales with reduced computational complexity. In turn, the discriminator network of the adversarial framework is formulated by combining convolutional layers with an additional squeeze excitation block to differentiate the generated segmentation mask from its respective ground truth. Before feeding the images to the network, we pre-processed them by using edge sharpening and Gaussian regularization to reach an optimized solution for vessel segmentation. The output of the trained model is post-processed using morphological operations to remove the small speckles of noise. The proposed method qualitatively and quantitatively outperforms state-of-the-art vessel segmentation methods using DRIVE and STARE datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge