Carla Sendra-Balcells
Generalisability of deep learning models in low-resource imaging settings: A fetal ultrasound study in 5 African countries
Sep 20, 2022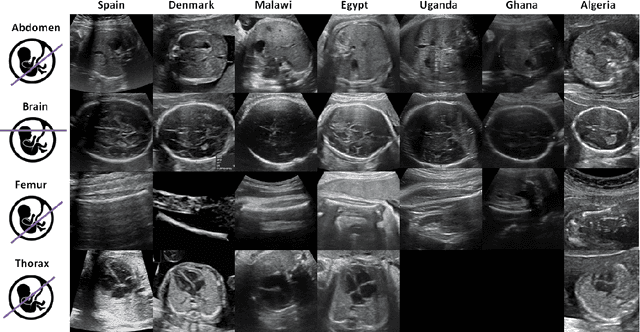
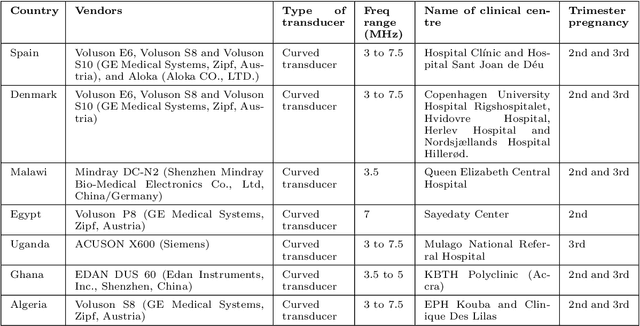
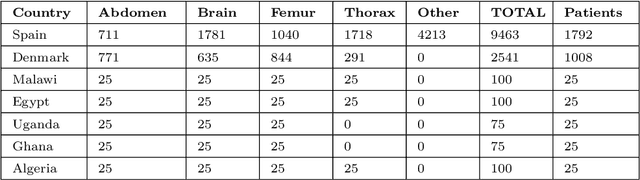
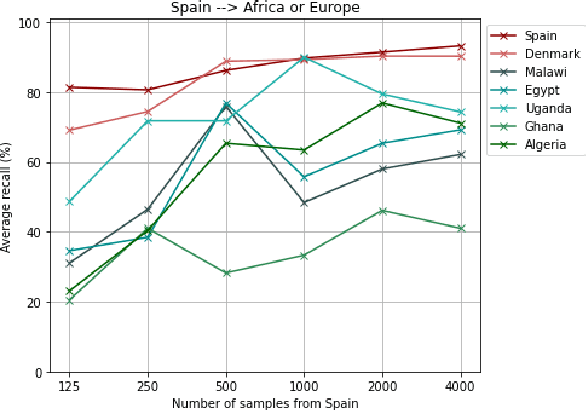
Abstract:Most artificial intelligence (AI) research have concentrated in high-income countries, where imaging data, IT infrastructures and clinical expertise are plentiful. However, slower progress has been made in limited-resource environments where medical imaging is needed. For example, in Sub-Saharan Africa the rate of perinatal mortality is very high due to limited access to antenatal screening. In these countries, AI models could be implemented to help clinicians acquire fetal ultrasound planes for diagnosis of fetal abnormalities. So far, deep learning models have been proposed to identify standard fetal planes, but there is no evidence of their ability to generalise in centres with limited access to high-end ultrasound equipment and data. This work investigates different strategies to reduce the domain-shift effect for a fetal plane classification model trained on a high-resource clinical centre and transferred to a new low-resource centre. To that end, a classifier trained with 1,792 patients from Spain is first evaluated on a new centre in Denmark in optimal conditions with 1,008 patients and is later optimised to reach the same performance in five African centres (Egypt, Algeria, Uganda, Ghana and Malawi) with 25 patients each. The results show that a transfer learning approach can be a solution to integrate small-size African samples with existing large-scale databases in developed countries. In particular, the model can be re-aligned and optimised to boost the performance on African populations by increasing the recall to $0.92 \pm 0.04$ and at the same time maintaining a high precision across centres. This framework shows promise for building new AI models generalisable across clinical centres with limited data acquired in challenging and heterogeneous conditions and calls for further research to develop new solutions for usability of AI in countries with less resources.
Multi-center, multi-vendor automated segmentation of left ventricular anatomy in contrast-enhanced MRI
Oct 28, 2021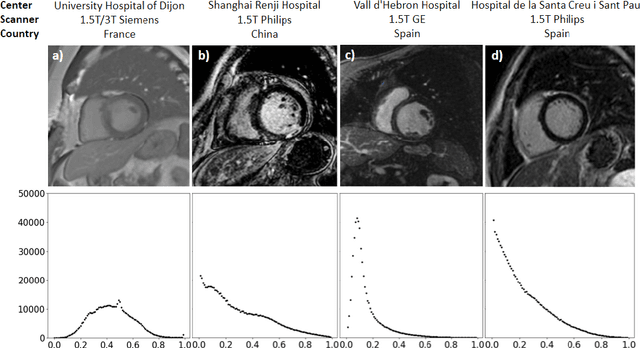
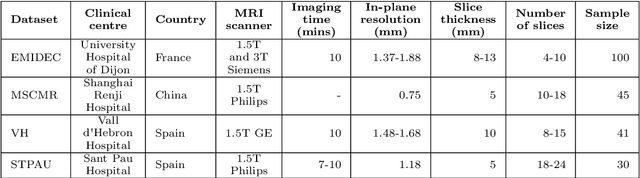
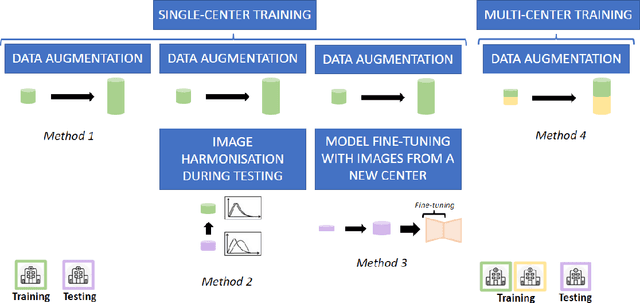
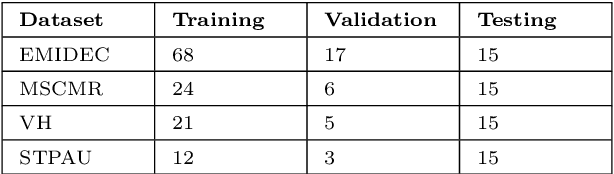
Abstract:Accurate delineation of the left ventricular boundaries in late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) is an essential step for scar tissue quantification and patient-specific assessment of myocardial infarction. Many deep-learning techniques have been proposed to perform automatic segmentations of the left ventricle (LV) in LGE-MRI showing segmentations as accurate as those obtained by expert cardiologists. Thus far, the existing models have been overwhelmingly developed and evaluated with LGE-MRI datasets from single clinical centers. However, in practice, LGE-MRI images vary significantly between clinical centers within and across countries, in particular due to differences in the MRI scanners, imaging conditions, contrast injection protocols and local clinical practise. This work investigates for the first time multi-center and multi-vendor LV segmentation in LGE-MRI, by proposing, implementing and evaluating in detail several strategies to enhance model generalizability across clinical cites. These include data augmentation to artificially augment the image variability in the training sample, image harmonization to align the distributions of LGE-MRI images across centers, and transfer learning to adjust existing single-center models to unseen images from new clinical sites. The results obtained based on a new multi-center LGE-MRI dataset acquired in four clinical centers in Spain, France and China, show that the combination of data augmentation and transfer learning can lead to single-center models that generalize well to new clinical centers not included in the original training. The proposed framework shows the potential for developing clinical tools for automated LV segmentation in LGE-MRI that can be deployed in multiple clinical centers across distinct geographical locations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge