Mathias Perslev
Fully Automated Tumor Segmentation for Brain MRI data using Multiplanner UNet
Jan 12, 2024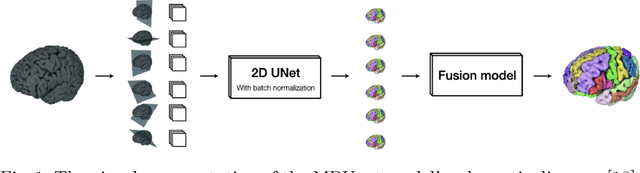
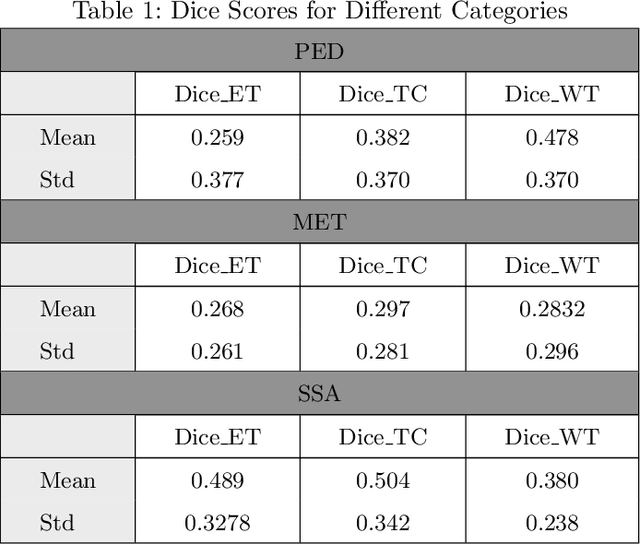
Abstract:Automated segmentation of distinct tumor regions is critical for accurate diagnosis and treatment planning in pediatric brain tumors. This study evaluates the efficacy of the Multi-Planner U-Net (MPUnet) approach in segmenting different tumor subregions across three challenging datasets: Pediatrics Tumor Challenge (PED), Brain Metastasis Challenge (MET), and Sub-Sahara-Africa Adult Glioma (SSA). These datasets represent diverse scenarios and anatomical variations, making them suitable for assessing the robustness and generalization capabilities of the MPUnet model. By utilizing multi-planar information, the MPUnet architecture aims to enhance segmentation accuracy. Our results show varying performance levels across the evaluated challenges, with the tumor core (TC) class demonstrating relatively higher segmentation accuracy. However, variability is observed in the segmentation of other classes, such as the edema and enhancing tumor (ET) regions. These findings emphasize the complexity of brain tumor segmentation and highlight the potential for further refinement of the MPUnet approach and inclusion of MRI more data and preprocessing.
Local Gamma Augmentation for Ischemic Stroke Lesion Segmentation on MRI
Jan 12, 2024Abstract:The identification and localisation of pathological tissues in medical images continues to command much attention among deep learning practitioners. When trained on abundant datasets, deep neural networks can match or exceed human performance. However, the scarcity of annotated data complicates the training of these models. Data augmentation techniques can compensate for a lack of training samples. However, many commonly used augmentation methods can fail to provide meaningful samples during model fitting. We present local gamma augmentation, a technique for introducing new instances of intensities in pathological tissues. We leverage local gamma augmentation to compensate for a bias in intensities corresponding to ischemic stroke lesions in human brain MRIs. On three datasets, we show how local gamma augmentation can improve the image-level sensitivity of a deep neural network tasked with ischemic lesion segmentation on magnetic resonance images.
The Medical Segmentation Decathlon
Jun 10, 2021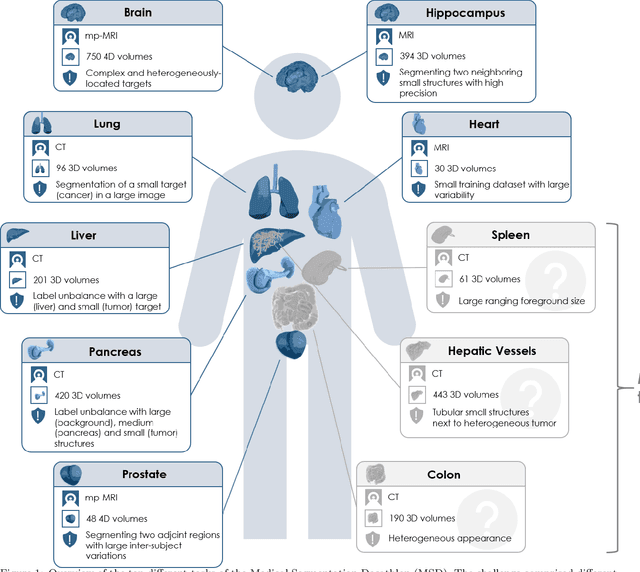
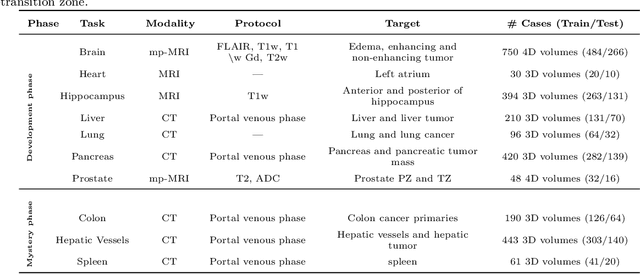
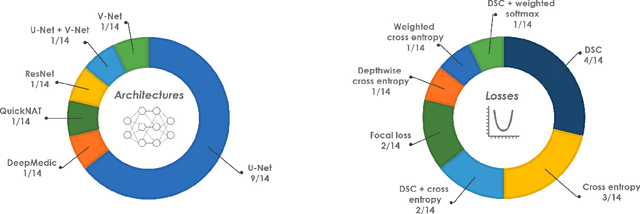
Abstract:International challenges have become the de facto standard for comparative assessment of image analysis algorithms given a specific task. Segmentation is so far the most widely investigated medical image processing task, but the various segmentation challenges have typically been organized in isolation, such that algorithm development was driven by the need to tackle a single specific clinical problem. We hypothesized that a method capable of performing well on multiple tasks will generalize well to a previously unseen task and potentially outperform a custom-designed solution. To investigate the hypothesis, we organized the Medical Segmentation Decathlon (MSD) - a biomedical image analysis challenge, in which algorithms compete in a multitude of both tasks and modalities. The underlying data set was designed to explore the axis of difficulties typically encountered when dealing with medical images, such as small data sets, unbalanced labels, multi-site data and small objects. The MSD challenge confirmed that algorithms with a consistent good performance on a set of tasks preserved their good average performance on a different set of previously unseen tasks. Moreover, by monitoring the MSD winner for two years, we found that this algorithm continued generalizing well to a wide range of other clinical problems, further confirming our hypothesis. Three main conclusions can be drawn from this study: (1) state-of-the-art image segmentation algorithms are mature, accurate, and generalize well when retrained on unseen tasks; (2) consistent algorithmic performance across multiple tasks is a strong surrogate of algorithmic generalizability; (3) the training of accurate AI segmentation models is now commoditized to non AI experts.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020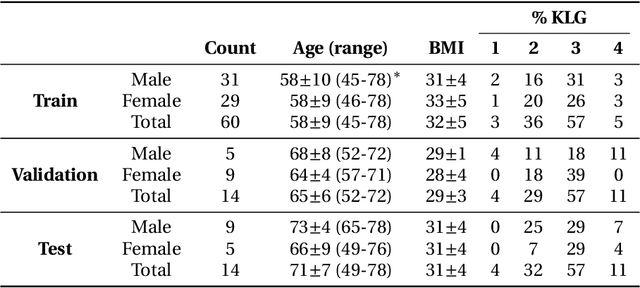
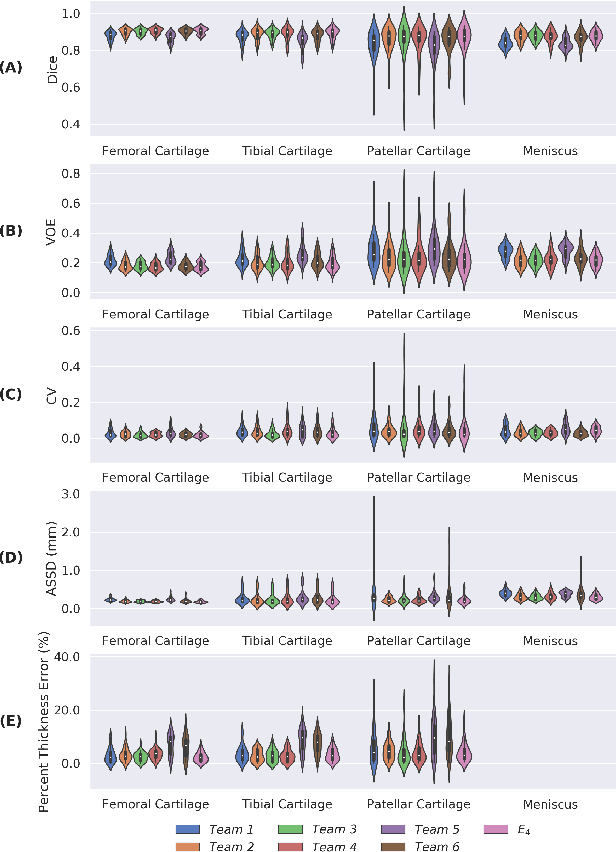
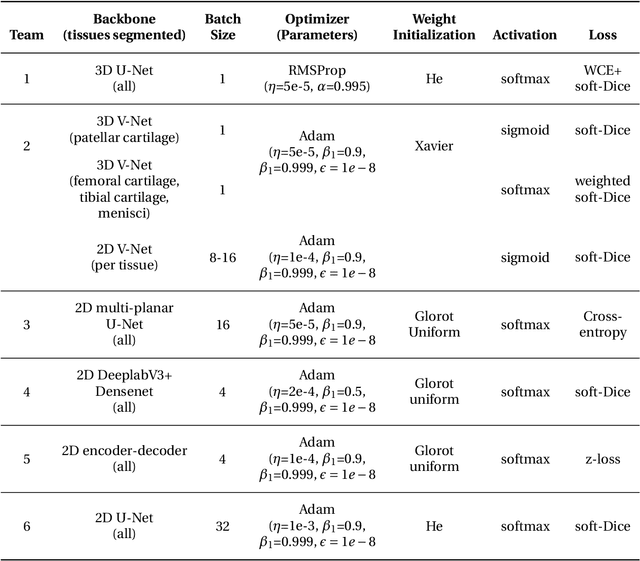
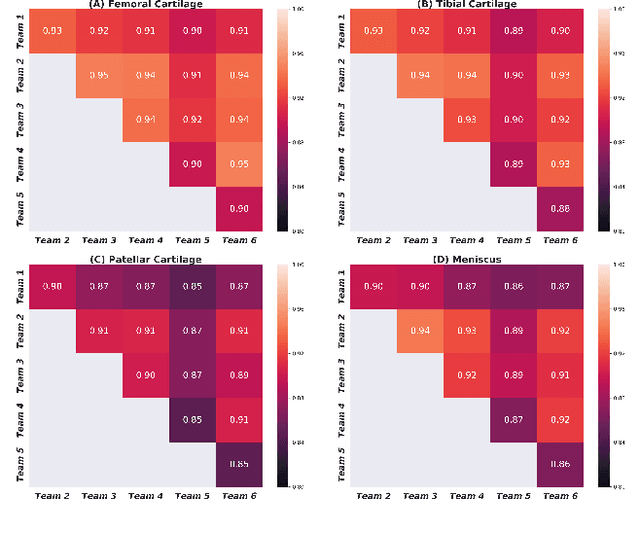
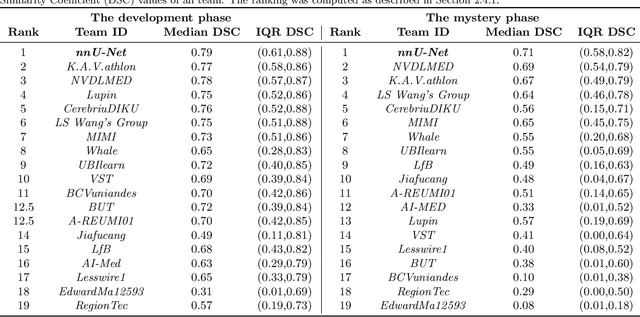
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
One Network to Segment Them All: A General, Lightweight System for Accurate 3D Medical Image Segmentation
Nov 05, 2019


Abstract:Many recent medical segmentation systems rely on powerful deep learning models to solve highly specific tasks. To maximize performance, it is standard practice to evaluate numerous pipelines with varying model topologies, optimization parameters, pre- & postprocessing steps, and even model cascades. It is often not clear how the resulting pipeline transfers to different tasks. We propose a simple and thoroughly evaluated deep learning framework for segmentation of arbitrary medical image volumes. The system requires no task-specific information, no human interaction and is based on a fixed model topology and a fixed hyperparameter set, eliminating the process of model selection and its inherent tendency to cause method-level over-fitting. The system is available in open source and does not require deep learning expertise to use. Without task-specific modifications, the system performed better than or similar to highly specialized deep learning methods across 3 separate segmentation tasks. In addition, it ranked 5-th and 6-th in the first and second round of the 2018 Medical Segmentation Decathlon comprising another 10 tasks. The system relies on multi-planar data augmentation which facilitates the application of a single 2D architecture based on the familiar U-Net. Multi-planar training combines the parameter efficiency of a 2D fully convolutional neural network with a systematic train- and test-time augmentation scheme, which allows the 2D model to learn a representation of the 3D image volume that fosters generalization.
U-Time: A Fully Convolutional Network for Time Series Segmentation Applied to Sleep Staging
Oct 24, 2019



Abstract:Neural networks are becoming more and more popular for the analysis of physiological time-series. The most successful deep learning systems in this domain combine convolutional and recurrent layers to extract useful features to model temporal relations. Unfortunately, these recurrent models are difficult to tune and optimize. In our experience, they often require task-specific modifications, which makes them challenging to use for non-experts. We propose U-Time, a fully feed-forward deep learning approach to physiological time series segmentation developed for the analysis of sleep data. U-Time is a temporal fully convolutional network based on the U-Net architecture that was originally proposed for image segmentation. U-Time maps sequential inputs of arbitrary length to sequences of class labels on a freely chosen temporal scale. This is done by implicitly classifying every individual time-point of the input signal and aggregating these classifications over fixed intervals to form the final predictions. We evaluated U-Time for sleep stage classification on a large collection of sleep electroencephalography (EEG) datasets. In all cases, we found that U-Time reaches or outperforms current state-of-the-art deep learning models while being much more robust in the training process and without requiring architecture or hyperparameter adaptation across tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge