Erik B Dam
Equity through Access: A Case for Small-scale Deep Learning
Mar 19, 2024



Abstract:The recent advances in deep learning (DL) have been accelerated by access to large-scale data and compute. These large-scale resources have been used to train progressively larger models which are resource intensive in terms of compute, data, energy, and carbon emissions. These costs are becoming a new type of entry barrier to researchers and practitioners with limited access to resources at such scale, particularly in the Global South. In this work, we take a comprehensive look at the landscape of existing DL models for vision tasks and demonstrate their usefulness in settings where resources are limited. To account for the resource consumption of DL models, we introduce a novel measure to estimate the performance per resource unit, which we call the PePR score. Using a diverse family of 131 unique DL architectures (spanning 1M to 130M trainable parameters) and three medical image datasets, we capture trends about the performance-resource trade-offs. In applications like medical image analysis, we argue that small-scale, specialized models are better than striving for large-scale models. Furthermore, we show that using pretrained models can significantly reduce the computational resources and data required. We hope this work will encourage the community to focus on improving AI equity by developing methods and models with smaller resource footprints.
Fully Automated Tumor Segmentation for Brain MRI data using Multiplanner UNet
Jan 12, 2024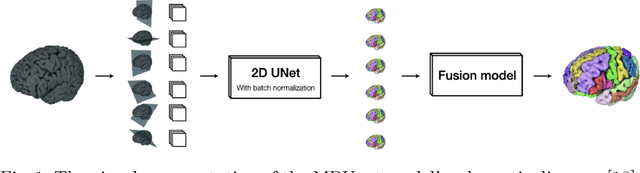
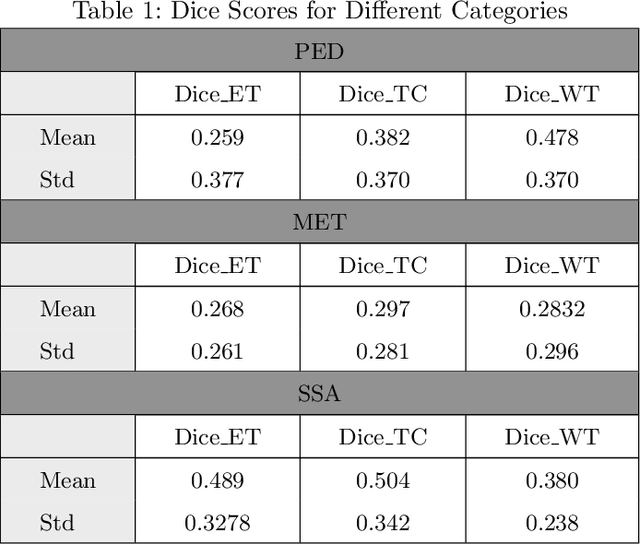
Abstract:Automated segmentation of distinct tumor regions is critical for accurate diagnosis and treatment planning in pediatric brain tumors. This study evaluates the efficacy of the Multi-Planner U-Net (MPUnet) approach in segmenting different tumor subregions across three challenging datasets: Pediatrics Tumor Challenge (PED), Brain Metastasis Challenge (MET), and Sub-Sahara-Africa Adult Glioma (SSA). These datasets represent diverse scenarios and anatomical variations, making them suitable for assessing the robustness and generalization capabilities of the MPUnet model. By utilizing multi-planar information, the MPUnet architecture aims to enhance segmentation accuracy. Our results show varying performance levels across the evaluated challenges, with the tumor core (TC) class demonstrating relatively higher segmentation accuracy. However, variability is observed in the segmentation of other classes, such as the edema and enhancing tumor (ET) regions. These findings emphasize the complexity of brain tumor segmentation and highlight the potential for further refinement of the MPUnet approach and inclusion of MRI more data and preprocessing.
Operating critical machine learning models in resource constrained regimes
Mar 17, 2023Abstract:The accelerated development of machine learning methods, primarily deep learning, are causal to the recent breakthroughs in medical image analysis and computer aided intervention. The resource consumption of deep learning models in terms of amount of training data, compute and energy costs are known to be massive. These large resource costs can be barriers in deploying these models in clinics, globally. To address this, there are cogent efforts within the machine learning community to introduce notions of resource efficiency. For instance, using quantisation to alleviate memory consumption. While most of these methods are shown to reduce the resource utilisation, they could come at a cost in performance. In this work, we probe into the trade-off between resource consumption and performance, specifically, when dealing with models that are used in critical settings such as in clinics.
Patch-based medical image segmentation using Quantum Tensor Networks
Sep 15, 2021



Abstract:Tensor networks are efficient factorisations of high dimensional tensors into a network of lower order tensors. They have been most commonly used to model entanglement in quantum many-body systems and more recently are witnessing increased applications in supervised machine learning. In this work, we formulate image segmentation in a supervised setting with tensor networks. The key idea is to first lift the pixels in image patches to exponentially high dimensional feature spaces and using a linear decision hyper-plane to classify the input pixels into foreground and background classes. The high dimensional linear model itself is approximated using the matrix product state (MPS) tensor network. The MPS is weight-shared between the non-overlapping image patches resulting in our strided tensor network model. The performance of the proposed model is evaluated on three 2D- and one 3D- biomedical imaging datasets. The performance of the proposed tensor network segmentation model is compared with relevant baseline methods. In the 2D experiments, the tensor network model yeilds competitive performance compared to the baseline methods while being more resource efficient.
Segmenting two-dimensional structures with strided tensor networks
Feb 13, 2021



Abstract:Tensor networks provide an efficient approximation of operations involving high dimensional tensors and have been extensively used in modelling quantum many-body systems. More recently, supervised learning has been attempted with tensor networks, primarily focused on tasks such as image classification. In this work, we propose a novel formulation of tensor networks for supervised image segmentation which allows them to operate on high resolution medical images. We use the matrix product state (MPS) tensor network on non-overlapping patches of a given input image to predict the segmentation mask by learning a pixel-wise linear classification rule in a high dimensional space. The proposed model is end-to-end trainable using backpropagation. It is implemented as a Strided Tensor Network to reduce the parameter complexity. The performance of the proposed method is evaluated on two public medical imaging datasets and compared to relevant baselines. The evaluation shows that the strided tensor network yields competitive performance compared to CNN-based models while using fewer resources. Additionally, based on the experiments we discuss the feasibility of using fully linear models for segmentation tasks.
Multi-layered tensor networks for image classification
Nov 13, 2020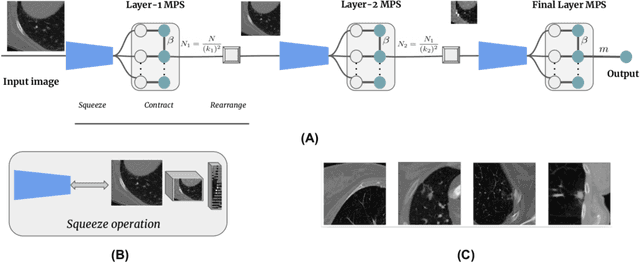

Abstract:The recently introduced locally orderless tensor network (LoTeNet) for supervised image classification uses matrix product state (MPS) operations on grids of transformed image patches. The resulting patch representations are combined back together into the image space and aggregated hierarchically using multiple MPS blocks per layer to obtain the final decision rules. In this work, we propose a non-patch based modification to LoTeNet that performs one MPS operation per layer, instead of several patch-level operations. The spatial information in the input images to MPS blocks at each layer is squeezed into the feature dimension, similar to LoTeNet, to maximise retained spatial correlation between pixels when images are flattened into 1D vectors. The proposed multi-layered tensor network (MLTN) is capable of learning linear decision boundaries in high dimensional spaces in a multi-layered setting, which results in a reduction in the computation cost compared to LoTeNet without any degradation in performance.
Locally orderless tensor networks for classifying two- and three-dimensional medical images
Sep 25, 2020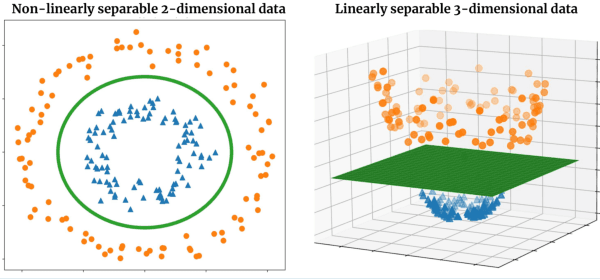
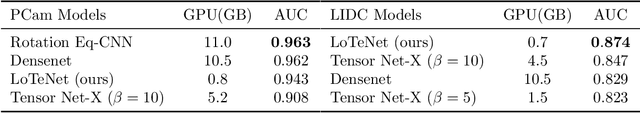
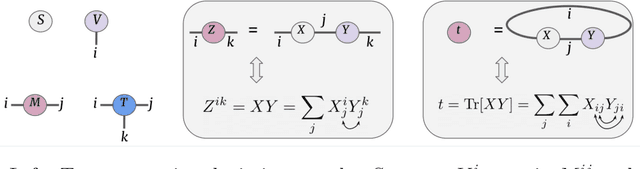
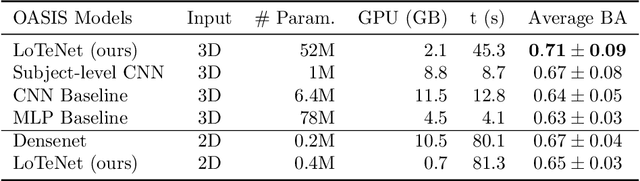
Abstract:Tensor networks are factorisations of high rank tensors into networks of lower rank tensors and have primarily been used to analyse quantum many-body problems. Tensor networks have seen a recent surge of interest in relation to supervised learning tasks with a focus on image classification. In this work, we improve upon the matrix product state (MPS) tensor networks that can operate on one-dimensional vectors to be useful for working with 2D and 3D medical images. We treat small image regions as orderless, squeeze their spatial information into feature dimensions and then perform MPS operations on these locally orderless regions. These local representations are then aggregated in a hierarchical manner to retain global structure. The proposed locally orderless tensor network (LoTeNet) is compared with relevant methods on three datasets. The architecture of LoTeNet is fixed in all experiments and we show it requires lesser computational resources to attain performance on par or superior to the compared methods.
Tensor Networks for Medical Image Classification
Apr 21, 2020



Abstract:With the increasing adoption of machine learning tools like neural networks across several domains, interesting connections and comparisons to concepts from other domains are coming to light. In this work, we focus on the class of Tensor Networks, which has been a work horse for physicists in the last two decades to analyse quantum many-body systems. Building on the recent interest in tensor networks for machine learning, we extend the Matrix Product State tensor networks (which can be interpreted as linear classifiers operating in exponentially high dimensional spaces) to be useful in medical image analysis tasks. We focus on classification problems as a first step where we motivate the use of tensor networks and propose adaptions for 2D images using classical image domain concepts such as local orderlessness of images. With the proposed locally orderless tensor network model (LoTeNet), we show that tensor networks are capable of attaining performance that is comparable to state-of-the-art deep learning methods. We evaluate the model on two publicly available medical imaging datasets and show performance improvements with fewer model hyperparameters and lesser computational resources compared to relevant baseline methods.
Simple Methods for Scanner Drift Normalization Validated for Automatic Segmentation of Knee Magnetic Resonance Imaging - with data from the Osteoarthritis Initiative
Dec 22, 2017



Abstract:Scanner drift is a well-known magnetic resonance imaging (MRI) artifact characterized by gradual signal degradation and scan intensity changes over time. In addition, hardware and software updates may imply abrupt changes in signal. The combined effects are particularly challenging for automatic image analysis methods used in longitudinal studies. The implication is increased measurement variation and a risk of bias in the estimations (e.g. in the volume change for a structure). We proposed two quite different approaches for scanner drift normalization and demonstrated the performance for segmentation of knee MRI using the fully automatic KneeIQ framework. The validation included a total of 1975 scans from both high-field and low-field MRI. The results demonstrated that the pre-processing method denoted Atlas Affine Normalization significantly removed scanner drift effects and ensured that the cartilage volume change quantifications became consistent with manual expert scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge