Geert Litjens
Democratizing Pathology Co-Pilots: An Open Pipeline and Dataset for Whole-Slide Vision-Language Modelling
Dec 19, 2025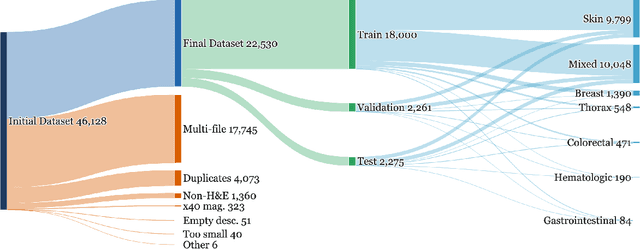
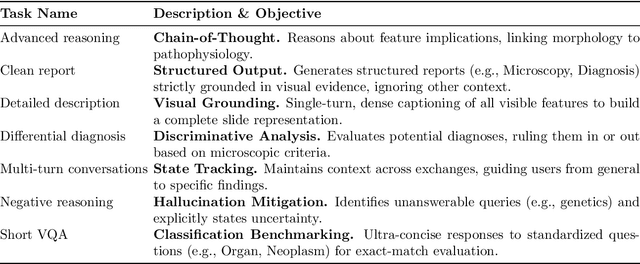
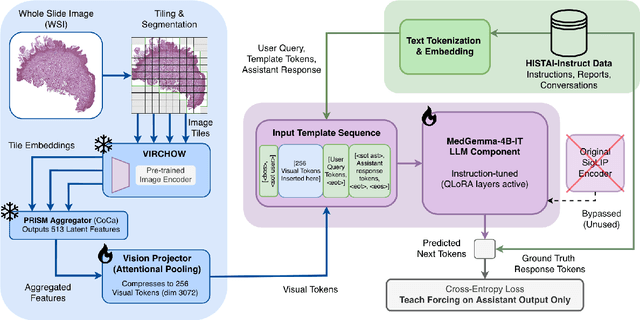
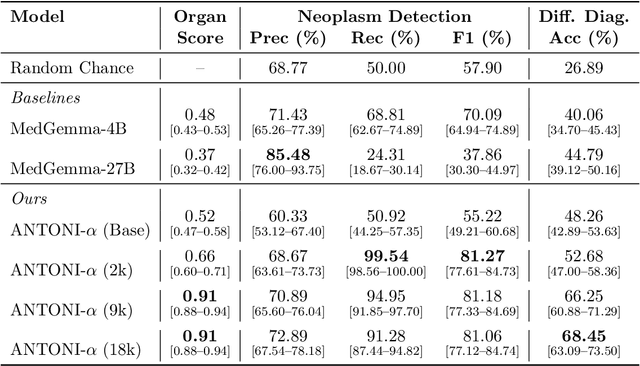
Abstract:Vision-language models (VLMs) have the potential to become co-pilots for pathologists. However, most VLMs either focus on small regions of interest within whole-slide images, provide only static slide-level outputs, or rely on data that is not publicly available, limiting reproducibility. Furthermore, training data containing WSIs paired with detailed clinical reports is scarce, restricting progress toward transparent and generalisable VLMs. We address these limitations with three main contributions. First, we introduce Polysome, a standardised tool for synthetic instruction generation. Second, we apply Polysome to the public HISTAI dataset, generating HISTAI-Instruct, a large whole-slide instruction tuning dataset spanning 24,259 slides and over 1.1 million instruction-response pairs. Finally, we use HISTAI-Instruct to train ANTONI-α, a VLM capable of visual-question answering (VQA). We show that ANTONI-α outperforms MedGemma on WSI-level VQA tasks of tissue identification, neoplasm detection, and differential diagnosis. We also compare the performance of multiple incarnations of ANTONI-α trained with different amounts of data. All methods, data, and code are publicly available.
Prototype-Based Multiple Instance Learning for Gigapixel Whole Slide Image Classification
Mar 11, 2025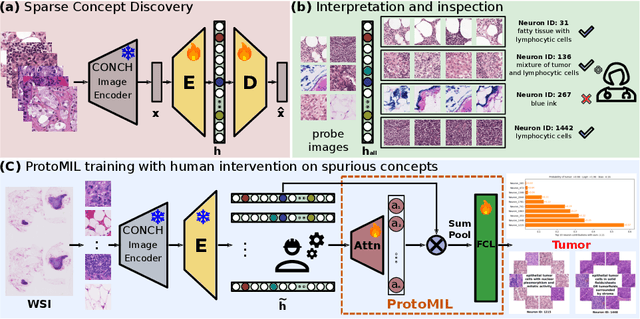
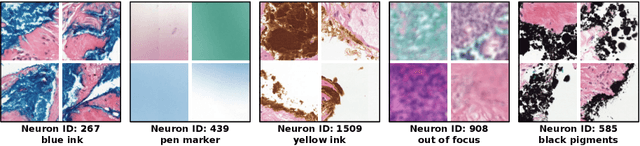
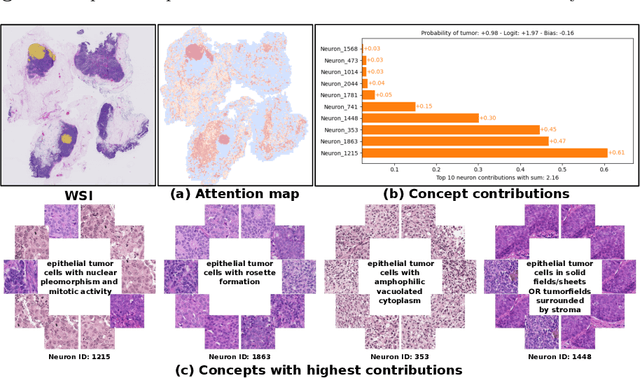
Abstract:Multiple Instance Learning (MIL) methods have succeeded remarkably in histopathology whole slide image (WSI) analysis. However, most MIL models only offer attention-based explanations that do not faithfully capture the model's decision mechanism and do not allow human-model interaction. To address these limitations, we introduce ProtoMIL, an inherently interpretable MIL model for WSI analysis that offers user-friendly explanations and supports human intervention. Our approach employs a sparse autoencoder to discover human-interpretable concepts from the image feature space, which are then used to train ProtoMIL. The model represents predictions as linear combinations of concepts, making the decision process transparent. Furthermore, ProtoMIL allows users to perform model interventions by altering the input concepts. Experiments on two widely used pathology datasets demonstrate that ProtoMIL achieves a classification performance comparable to state-of-the-art MIL models while offering intuitively understandable explanations. Moreover, we demonstrate that our method can eliminate reliance on diagnostically irrelevant information via human intervention, guiding the model toward being right for the right reason. Code will be publicly available at https://github.com/ss-sun/ProtoMIL.
Label-free Concept Based Multiple Instance Learning for Gigapixel Histopathology
Jan 06, 2025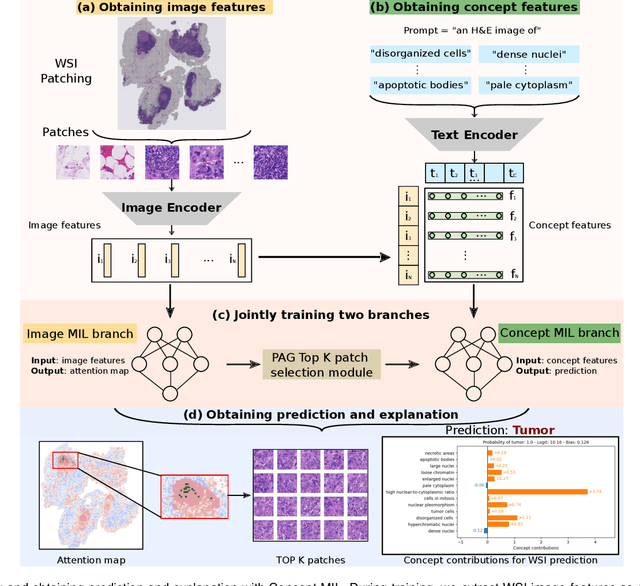
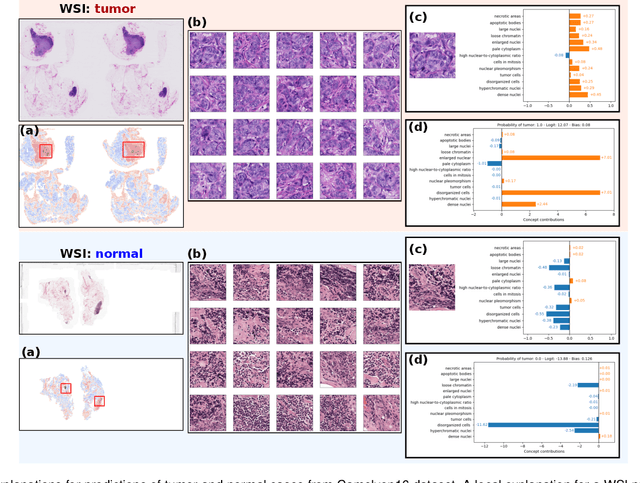
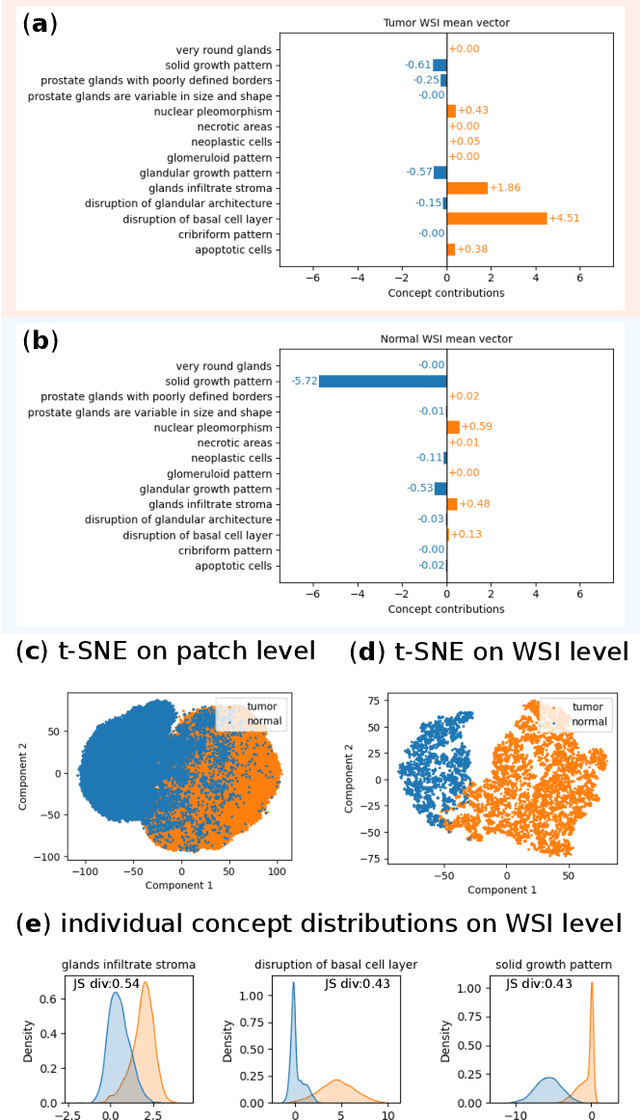
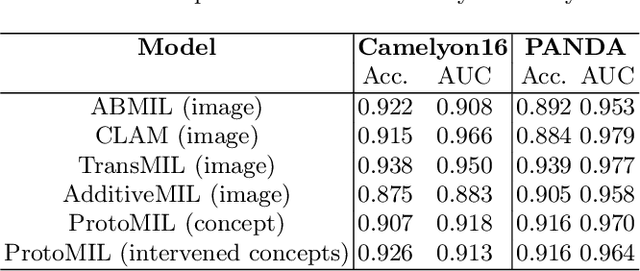
Abstract:Multiple Instance Learning (MIL) methods allow for gigapixel Whole-Slide Image (WSI) analysis with only slide-level annotations. Interpretability is crucial for safely deploying such algorithms in high-stakes medical domains. Traditional MIL methods offer explanations by highlighting salient regions. However, such spatial heatmaps provide limited insights for end users. To address this, we propose a novel inherently interpretable WSI-classification approach that uses human-understandable pathology concepts to generate explanations. Our proposed Concept MIL model leverages recent advances in vision-language models to directly predict pathology concepts based on image features. The model's predictions are obtained through a linear combination of the concepts identified on the top-K patches of a WSI, enabling inherent explanations by tracing each concept's influence on the prediction. In contrast to traditional concept-based interpretable models, our approach eliminates the need for costly human annotations by leveraging the vision-language model. We validate our method on two widely used pathology datasets: Camelyon16 and PANDA. On both datasets, Concept MIL achieves AUC and accuracy scores over 0.9, putting it on par with state-of-the-art models. We further find that 87.1\% (Camelyon16) and 85.3\% (PANDA) of the top 20 patches fall within the tumor region. A user study shows that the concepts identified by our model align with the concepts used by pathologists, making it a promising strategy for human-interpretable WSI classification.
Navigating the landscape of multimodal AI in medicine: a scoping review on technical challenges and clinical applications
Nov 06, 2024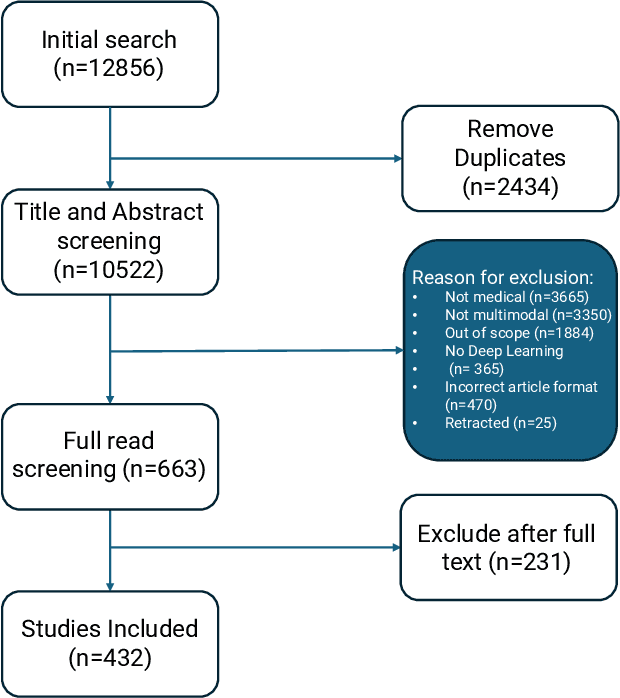
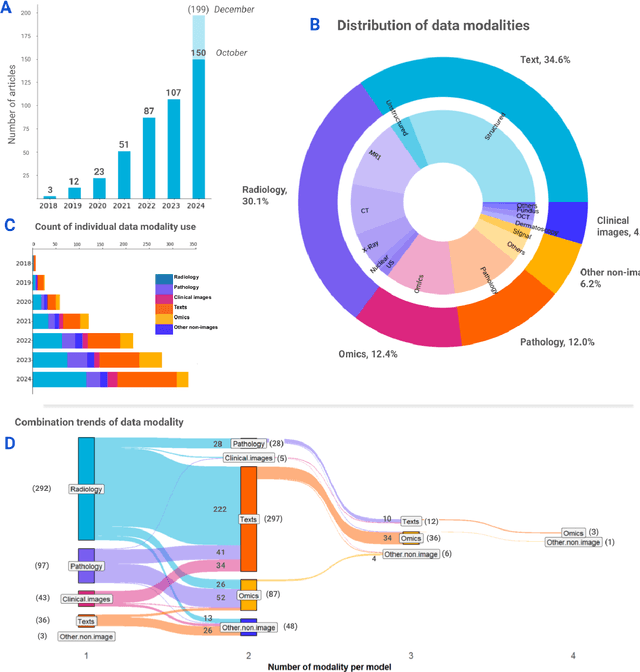
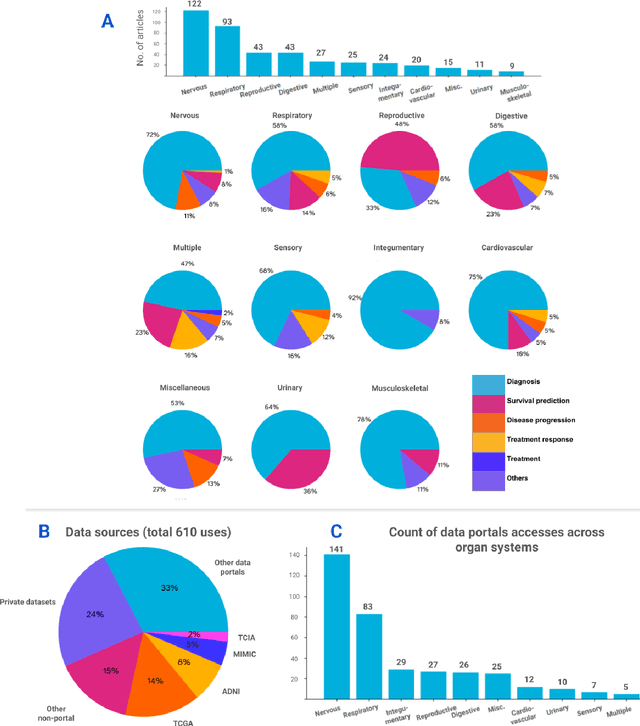
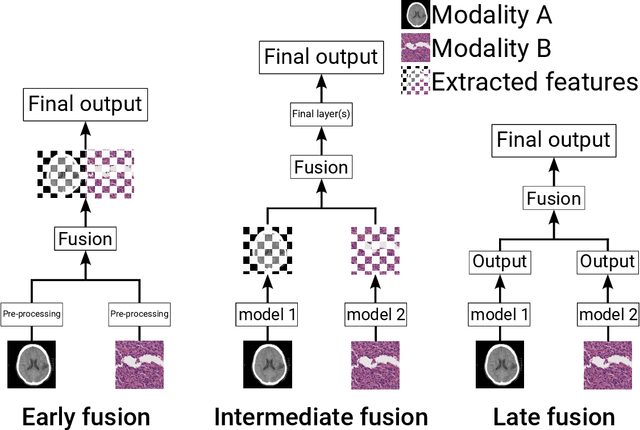
Abstract:Recent technological advances in healthcare have led to unprecedented growth in patient data quantity and diversity. While artificial intelligence (AI) models have shown promising results in analyzing individual data modalities, there is increasing recognition that models integrating multiple complementary data sources, so-called multimodal AI, could enhance clinical decision-making. This scoping review examines the landscape of deep learning-based multimodal AI applications across the medical domain, analyzing 432 papers published between 2018 and 2024. We provide an extensive overview of multimodal AI development across different medical disciplines, examining various architectural approaches, fusion strategies, and common application areas. Our analysis reveals that multimodal AI models consistently outperform their unimodal counterparts, with an average improvement of 6.2 percentage points in AUC. However, several challenges persist, including cross-departmental coordination, heterogeneous data characteristics, and incomplete datasets. We critically assess the technical and practical challenges in developing multimodal AI systems and discuss potential strategies for their clinical implementation, including a brief overview of commercially available multimodal AI models for clinical decision-making. Additionally, we identify key factors driving multimodal AI development and propose recommendations to accelerate the field's maturation. This review provides researchers and clinicians with a thorough understanding of the current state, challenges, and future directions of multimodal AI in medicine.
Masked Attention as a Mechanism for Improving Interpretability of Vision Transformers
Apr 28, 2024
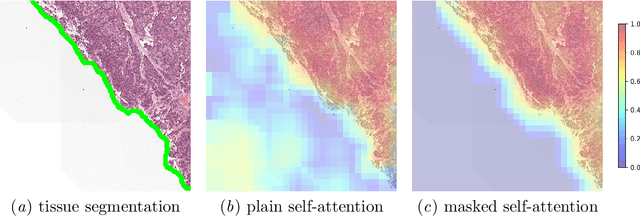
Abstract:Vision Transformers are at the heart of the current surge of interest in foundation models for histopathology. They process images by breaking them into smaller patches following a regular grid, regardless of their content. Yet, not all parts of an image are equally relevant for its understanding. This is particularly true in computational pathology where background is completely non-informative and may introduce artefacts that could mislead predictions. To address this issue, we propose a novel method that explicitly masks background in Vision Transformers' attention mechanism. This ensures tokens corresponding to background patches do not contribute to the final image representation, thereby improving model robustness and interpretability. We validate our approach using prostate cancer grading from whole-slide images as a case study. Our results demonstrate that it achieves comparable performance with plain self-attention while providing more accurate and clinically meaningful attention heatmaps.
Hierarchical Vision Transformers for Context-Aware Prostate Cancer Grading in Whole Slide Images
Dec 19, 2023Abstract:Vision Transformers (ViTs) have ushered in a new era in computer vision, showcasing unparalleled performance in many challenging tasks. However, their practical deployment in computational pathology has largely been constrained by the sheer size of whole slide images (WSIs), which result in lengthy input sequences. Transformers faced a similar limitation when applied to long documents, and Hierarchical Transformers were introduced to circumvent it. Given the analogous challenge with WSIs and their inherent hierarchical structure, Hierarchical Vision Transformers (H-ViTs) emerge as a promising solution in computational pathology. This work delves into the capabilities of H-ViTs, evaluating their efficiency for prostate cancer grading in WSIs. Our results show that they achieve competitive performance against existing state-of-the-art solutions.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
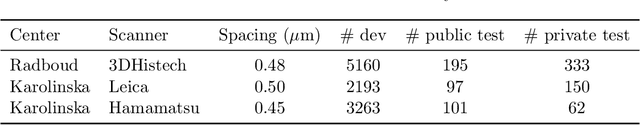
Domain adaptation strategies for cancer-independent detection of lymph node metastases
Jul 13, 2022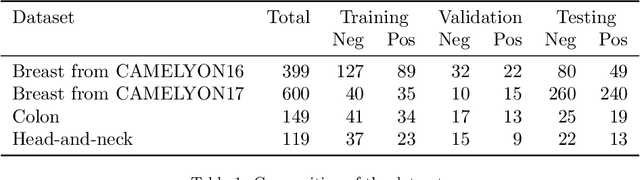

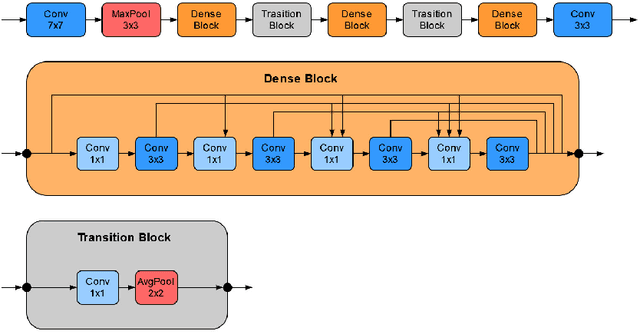
Abstract:Recently, large, high-quality public datasets have led to the development of convolutional neural networks that can detect lymph node metastases of breast cancer at the level of expert pathologists. Many cancers, regardless of the site of origin, can metastasize to lymph nodes. However, collecting and annotating high-volume, high-quality datasets for every cancer type is challenging. In this paper we investigate how to leverage existing high-quality datasets most efficiently in multi-task settings for closely related tasks. Specifically, we will explore different training and domain adaptation strategies, including prevention of catastrophic forgetting, for colon and head-and-neck cancer metastasis detection in lymph nodes. Our results show state-of-the-art performance on both cancer metastasis detection tasks. Furthermore, we show the effectiveness of repeated adaptation of networks from one cancer type to another to obtain multi-task metastasis detection networks. Last, we show that leveraging existing high-quality datasets can significantly boost performance on new target tasks and that catastrophic forgetting can be effectively mitigated using regularization.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
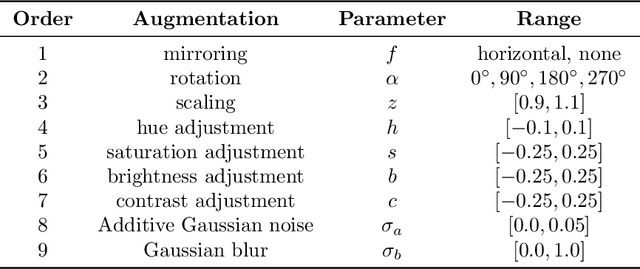
Jun 03, 2022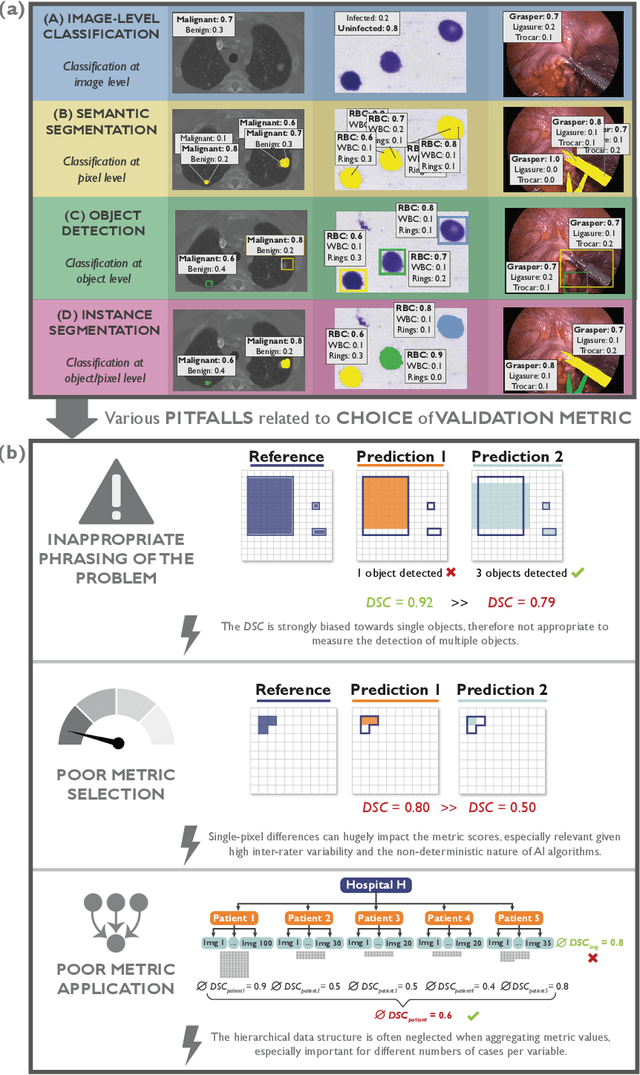
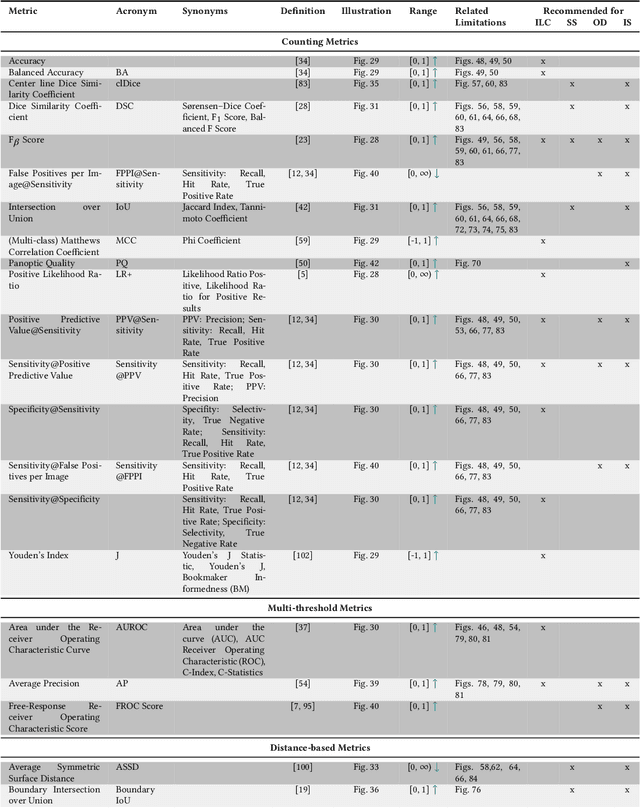
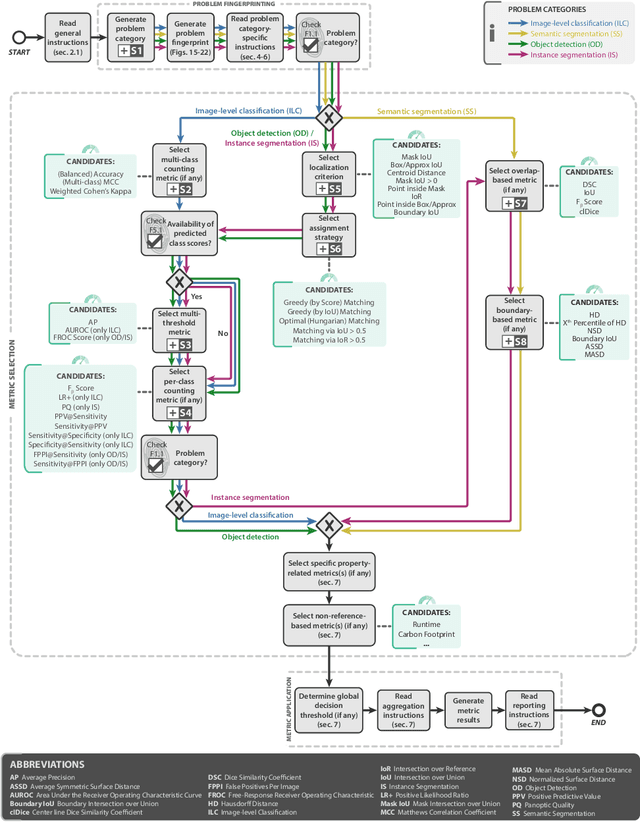
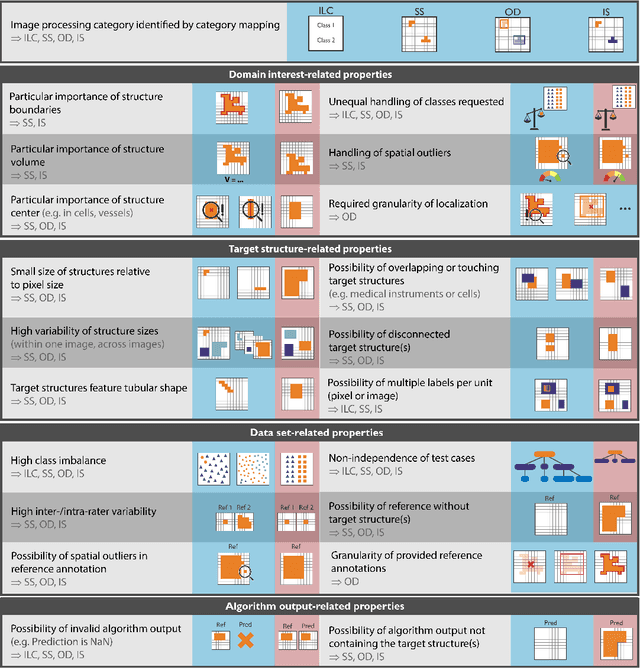
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
Automatic tumour segmentation in H&E-stained whole-slide images of the pancreas
Dec 01, 2021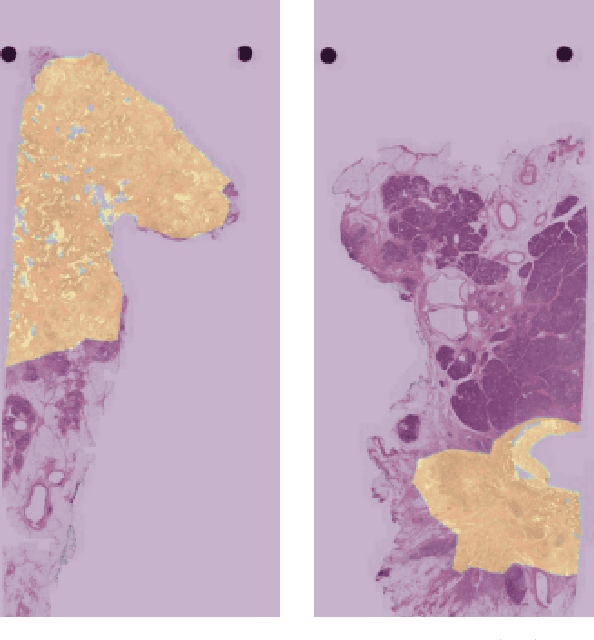
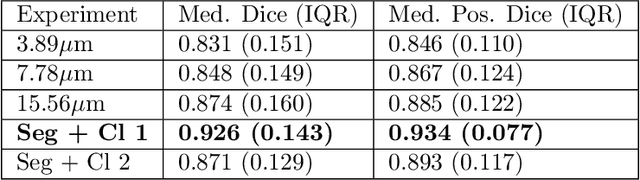
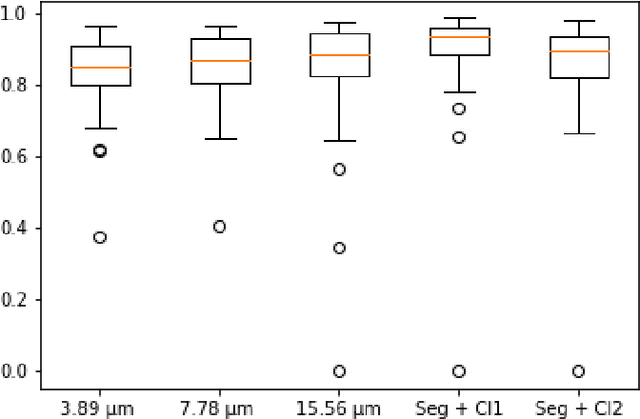
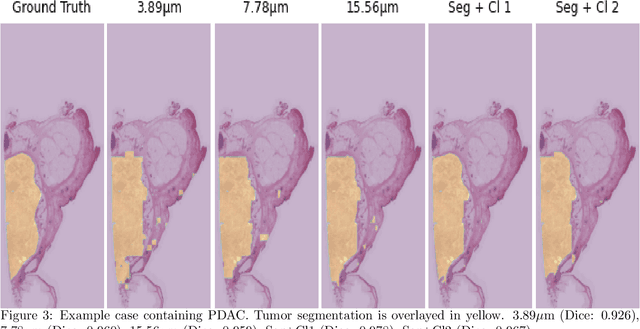
Abstract:Pancreatic cancer will soon be the second leading cause of cancer-related death in Western society. Imaging techniques such as CT, MRI and ultrasound typically help providing the initial diagnosis, but histopathological assessment is still the gold standard for final confirmation of disease presence and prognosis. In recent years machine learning approaches and pathomics pipelines have shown potential in improving diagnostics and prognostics in other cancerous entities, such as breast and prostate cancer. A crucial first step in these pipelines is typically identification and segmentation of the tumour area. Ideally this step is done automatically to prevent time consuming manual annotation. We propose a multi-task convolutional neural network to balance disease detection and segmentation accuracy. We validated our approach on a dataset of 29 patients (for a total of 58 slides) at different resolutions. The best single task segmentation network achieved a median Dice of 0.885 (0.122) IQR at a resolution of 15.56 $\mu$m. Our multi-task network improved on that with a median Dice score of 0.934 (0.077) IQR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge