Clément Grisi
Label-free Concept Based Multiple Instance Learning for Gigapixel Histopathology
Jan 06, 2025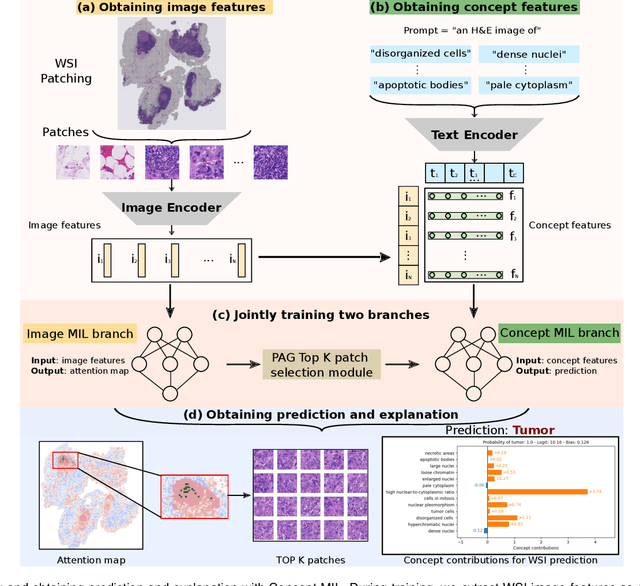
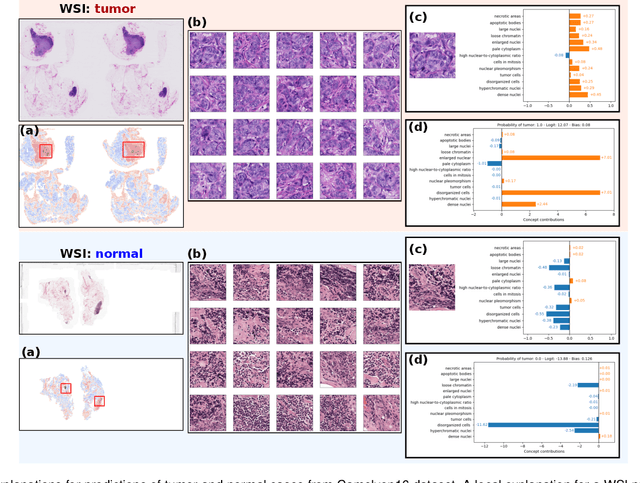
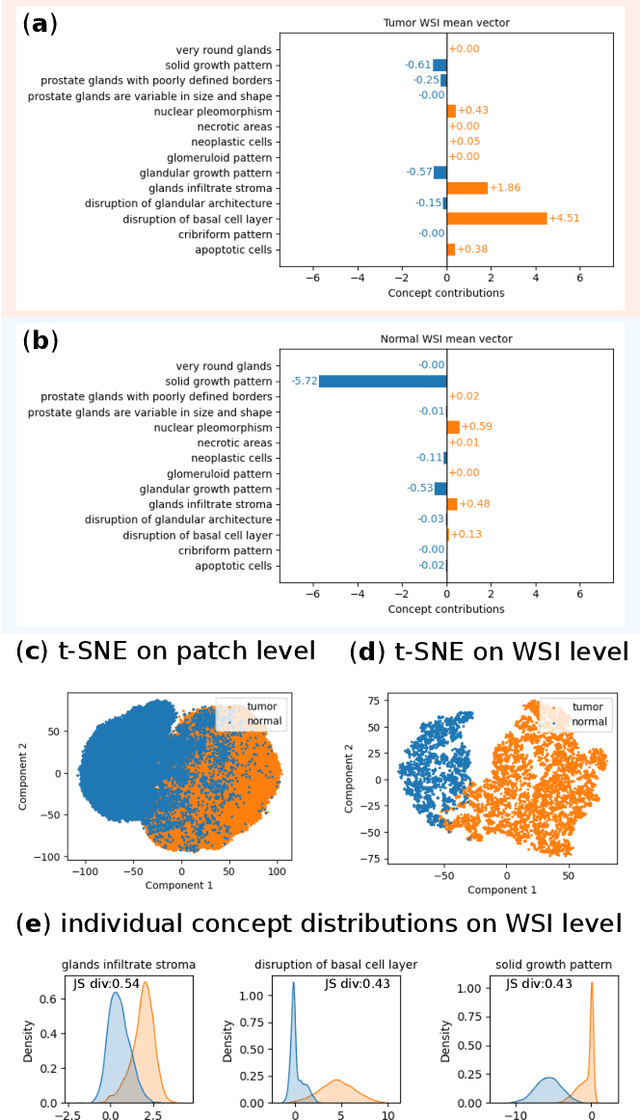
Abstract:Multiple Instance Learning (MIL) methods allow for gigapixel Whole-Slide Image (WSI) analysis with only slide-level annotations. Interpretability is crucial for safely deploying such algorithms in high-stakes medical domains. Traditional MIL methods offer explanations by highlighting salient regions. However, such spatial heatmaps provide limited insights for end users. To address this, we propose a novel inherently interpretable WSI-classification approach that uses human-understandable pathology concepts to generate explanations. Our proposed Concept MIL model leverages recent advances in vision-language models to directly predict pathology concepts based on image features. The model's predictions are obtained through a linear combination of the concepts identified on the top-K patches of a WSI, enabling inherent explanations by tracing each concept's influence on the prediction. In contrast to traditional concept-based interpretable models, our approach eliminates the need for costly human annotations by leveraging the vision-language model. We validate our method on two widely used pathology datasets: Camelyon16 and PANDA. On both datasets, Concept MIL achieves AUC and accuracy scores over 0.9, putting it on par with state-of-the-art models. We further find that 87.1\% (Camelyon16) and 85.3\% (PANDA) of the top 20 patches fall within the tumor region. A user study shows that the concepts identified by our model align with the concepts used by pathologists, making it a promising strategy for human-interpretable WSI classification.
Masked Attention as a Mechanism for Improving Interpretability of Vision Transformers
Apr 28, 2024
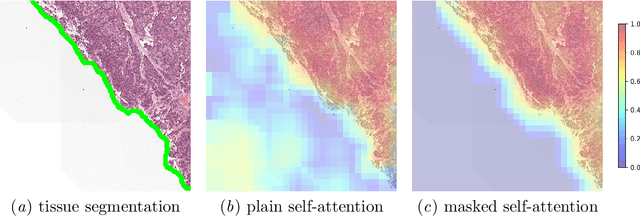
Abstract:Vision Transformers are at the heart of the current surge of interest in foundation models for histopathology. They process images by breaking them into smaller patches following a regular grid, regardless of their content. Yet, not all parts of an image are equally relevant for its understanding. This is particularly true in computational pathology where background is completely non-informative and may introduce artefacts that could mislead predictions. To address this issue, we propose a novel method that explicitly masks background in Vision Transformers' attention mechanism. This ensures tokens corresponding to background patches do not contribute to the final image representation, thereby improving model robustness and interpretability. We validate our approach using prostate cancer grading from whole-slide images as a case study. Our results demonstrate that it achieves comparable performance with plain self-attention while providing more accurate and clinically meaningful attention heatmaps.
Hierarchical Vision Transformers for Context-Aware Prostate Cancer Grading in Whole Slide Images
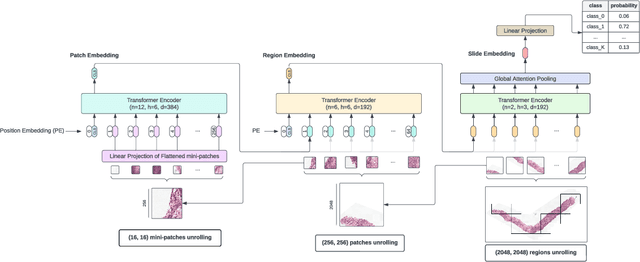
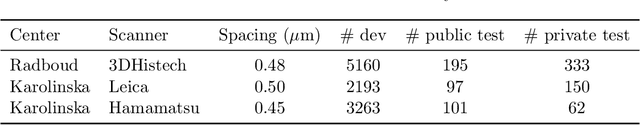
Dec 19, 2023Abstract:Vision Transformers (ViTs) have ushered in a new era in computer vision, showcasing unparalleled performance in many challenging tasks. However, their practical deployment in computational pathology has largely been constrained by the sheer size of whole slide images (WSIs), which result in lengthy input sequences. Transformers faced a similar limitation when applied to long documents, and Hierarchical Transformers were introduced to circumvent it. Given the analogous challenge with WSIs and their inherent hierarchical structure, Hierarchical Vision Transformers (H-ViTs) emerge as a promising solution in computational pathology. This work delves into the capabilities of H-ViTs, evaluating their efficiency for prostate cancer grading in WSIs. Our results show that they achieve competitive performance against existing state-of-the-art solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge