Erik Bjørnager Dam
deCIFer: Crystal Structure Prediction from Powder Diffraction Data using Autoregressive Language Models
Feb 04, 2025Abstract:Novel materials drive progress across applications from energy storage to electronics. Automated characterization of material structures with machine learning methods offers a promising strategy for accelerating this key step in material design. In this work, we introduce an autoregressive language model that performs crystal structure prediction (CSP) from powder diffraction data. The presented model, deCIFer, generates crystal structures in the widely used Crystallographic Information File (CIF) format and can be conditioned on powder X-ray diffraction (PXRD) data. Unlike earlier works that primarily rely on high-level descriptors like composition, deCIFer performs CSP from diffraction data. We train deCIFer on nearly 2.3M unique crystal structures and validate on diverse sets of PXRD patterns for characterizing challenging inorganic crystal systems. Qualitative and quantitative assessments using the residual weighted profile and Wasserstein distance show that deCIFer produces structures that more accurately match the target diffraction data when conditioned, compared to the unconditioned case. Notably, deCIFer can achieve a 94% match rate on unseen data. deCIFer bridges experimental diffraction data with computational CSP, lending itself as a powerful tool for crystal structure characterization and accelerating materials discovery.
UNet Architectures in Multiplanar Volumetric Segmentation -- Validated on Three Knee MRI Cohorts
Mar 15, 2022
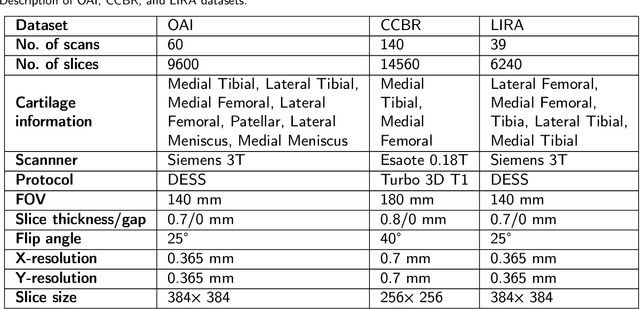

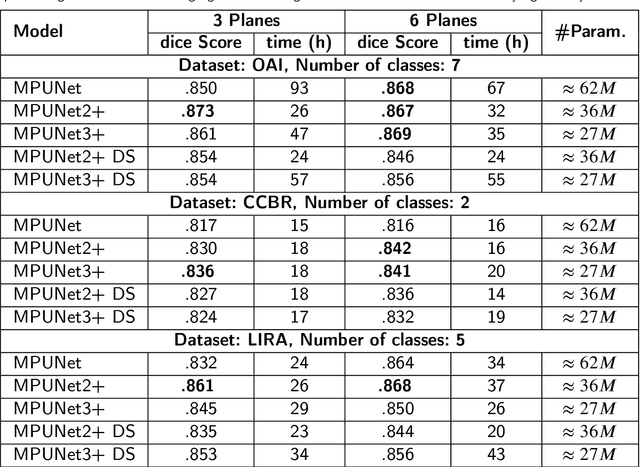
Abstract:UNet has become the gold standard method for segmenting 2D medical images that any new method must be validated against. However, in recent years, several variations of the seminal UNet have been proposed with promising results. However, there is no clear consensus on the generalisability of these architectures, and UNet currently remains the methodological gold standard. The purpose of this study was to evaluate some of the most promising UNet-inspired architectures for 3D segmentation. For the segmentation of 3D scans, UNet-inspired methods are also dominant, but there is a larger variety across applications. By evaluating the architectures in a different dimensionality, embedded in a different method, and for a different task, we aimed to evaluate if any of these UNet-alternatives are promising as a new gold standard that generalizes even better than UNet. Specifically, we investigated the architectures as the central 2D segmentation core in the Multi-Planar Unet 3D segmentation method that previously demonstrated excellent generalization in the MICCAI Segmentation Decathlon. Generalisability can be demonstrated if a promising UNet-variant consistently outperforms UNet in this setting. For this purpose, we evaluated four architectures for cartilage segmentation from three different cohorts with knee MRIs.
One Network to Segment Them All: A General, Lightweight System for Accurate 3D Medical Image Segmentation
Nov 05, 2019


Abstract:Many recent medical segmentation systems rely on powerful deep learning models to solve highly specific tasks. To maximize performance, it is standard practice to evaluate numerous pipelines with varying model topologies, optimization parameters, pre- & postprocessing steps, and even model cascades. It is often not clear how the resulting pipeline transfers to different tasks. We propose a simple and thoroughly evaluated deep learning framework for segmentation of arbitrary medical image volumes. The system requires no task-specific information, no human interaction and is based on a fixed model topology and a fixed hyperparameter set, eliminating the process of model selection and its inherent tendency to cause method-level over-fitting. The system is available in open source and does not require deep learning expertise to use. Without task-specific modifications, the system performed better than or similar to highly specialized deep learning methods across 3 separate segmentation tasks. In addition, it ranked 5-th and 6-th in the first and second round of the 2018 Medical Segmentation Decathlon comprising another 10 tasks. The system relies on multi-planar data augmentation which facilitates the application of a single 2D architecture based on the familiar U-Net. Multi-planar training combines the parameter efficiency of a 2D fully convolutional neural network with a systematic train- and test-time augmentation scheme, which allows the 2D model to learn a representation of the 3D image volume that fosters generalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge