Anders Bjorholm Dahl
Towards Agnostic and Holistic Universal Image Segmentation with Bit Diffusion
Jan 06, 2026Abstract:This paper introduces a diffusion-based framework for universal image segmentation, making agnostic segmentation possible without depending on mask-based frameworks and instead predicting the full segmentation in a holistic manner. We present several key adaptations to diffusion models, which are important in this discrete setting. Notably, we show that a location-aware palette with our 2D gray code ordering improves performance. Adding a final tanh activation function is crucial for discrete data. On optimizing diffusion parameters, the sigmoid loss weighting consistently outperforms alternatives, regardless of the prediction type used, and we settle on x-prediction. While our current model does not yet surpass leading mask-based architectures, it narrows the performance gap and introduces unique capabilities, such as principled ambiguity modeling, that these models lack. All models were trained from scratch, and we believe that combining our proposed improvements with large-scale pretraining or promptable conditioning could lead to competitive models.
Diffusion Based Ambiguous Image Segmentation
Apr 08, 2025Abstract:Medical image segmentation often involves inherent uncertainty due to variations in expert annotations. Capturing this uncertainty is an important goal and previous works have used various generative image models for the purpose of representing the full distribution of plausible expert ground truths. In this work, we explore the design space of diffusion models for generative segmentation, investigating the impact of noise schedules, prediction types, and loss weightings. Notably, we find that making the noise schedule harder with input scaling significantly improves performance. We conclude that x- and v-prediction outperform epsilon-prediction, likely because the diffusion process is in the discrete segmentation domain. Many loss weightings achieve similar performance as long as they give enough weight to the end of the diffusion process. We base our experiments on the LIDC-IDRI lung lesion dataset and obtain state-of-the-art (SOTA) performance. Additionally, we introduce a randomly cropped variant of the LIDC-IDRI dataset that is better suited for uncertainty in image segmentation. Our model also achieves SOTA in this harder setting.
Fast Sphericity and Roundness approximation in 2D and 3D using Local Thickness
Apr 08, 2025Abstract:Sphericity and roundness are fundamental measures used for assessing object uniformity in 2D and 3D images. However, using their strict definition makes computation costly. As both 2D and 3D microscopy imaging datasets grow larger, there is an increased demand for efficient algorithms that can quantify multiple objects in large volumes. We propose a novel approach for extracting sphericity and roundness based on the output of a local thickness algorithm. For sphericity, we simplify the surface area computation by modeling objects as spheroids/ellipses of varying lengths and widths of mean local thickness. For roundness, we avoid a complex corner curvature determination process by approximating it with local thickness values on the contour/surface of the object. The resulting methods provide an accurate representation of the exact measures while being significantly faster than their existing implementations.
Disconnect to Connect: A Data Augmentation Method for Improving Topology Accuracy in Image Segmentation
Mar 07, 2025Abstract:Accurate segmentation of thin, tubular structures (e.g., blood vessels) is challenging for deep neural networks. These networks classify individual pixels, and even minor misclassifications can break the thin connections within these structures. Existing methods for improving topology accuracy, such as topology loss functions, rely on very precise, topologically-accurate training labels, which are difficult to obtain. This is because annotating images, especially 3D images, is extremely laborious and time-consuming. Low image resolution and contrast further complicates the annotation by causing tubular structures to appear disconnected. We present CoLeTra, a data augmentation strategy that integrates to the models the prior knowledge that structures that appear broken are actually connected. This is achieved by creating images with the appearance of disconnected structures while maintaining the original labels. Our extensive experiments, involving different architectures, loss functions, and datasets, demonstrate that CoLeTra leads to segmentations topologically more accurate while often improving the Dice coefficient and Hausdorff distance. CoLeTra's hyper-parameters are intuitive to tune, and our sensitivity analysis shows that CoLeTra is robust to changes in these hyper-parameters. We also release a dataset specifically suited for image segmentation methods with a focus on topology accuracy. CoLetra's code can be found at https://github.com/jmlipman/CoLeTra.
TopoMortar: A dataset to evaluate image segmentation methods focused on topology accuracy
Mar 05, 2025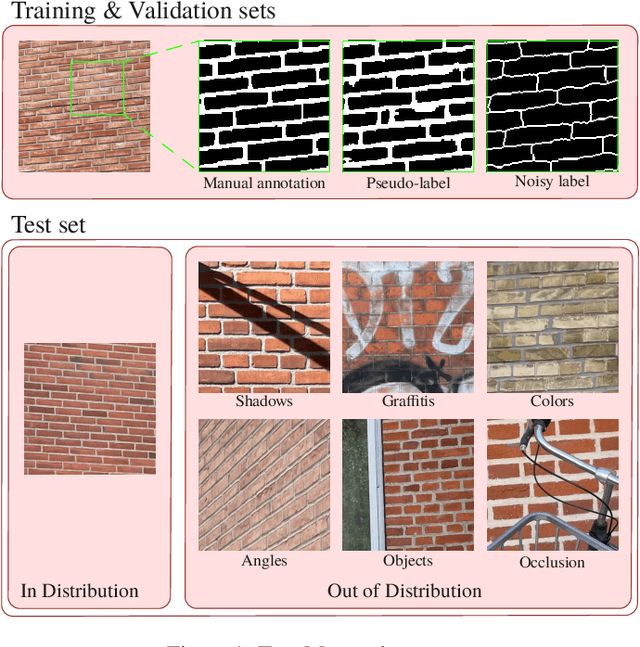
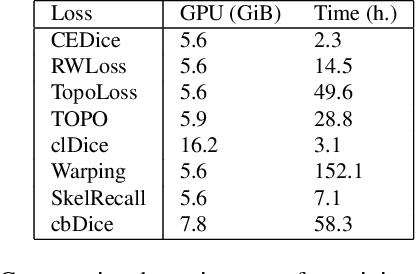
Abstract:We present TopoMortar, a brick wall dataset that is the first dataset specifically designed to evaluate topology-focused image segmentation methods, such as topology loss functions. TopoMortar enables to investigate in two ways whether methods incorporate prior topological knowledge. First, by eliminating challenges seen in real-world data, such as small training set, noisy labels, and out-of-distribution test-set images, that, as we show, impact the effectiveness of topology losses. Second, by allowing to assess in the same dataset topology accuracy across dataset challenges, isolating dataset-related effects from the effect of incorporating prior topological knowledge. In these two experiments, it is deliberately difficult to improve topology accuracy without actually using topology information, thus, permitting to attribute an improvement in topology accuracy to the incorporation of prior topological knowledge. To this end, TopoMortar includes three types of labels (accurate, noisy, pseudo-labels), two fixed training sets (large and small), and in-distribution and out-of-distribution test-set images. We compared eight loss functions on TopoMortar, and we found that clDice achieved the most topologically accurate segmentations, Skeleton Recall loss performed best particularly with noisy labels, and the relative advantageousness of the other loss functions depended on the experimental setting. Additionally, we show that simple methods, such as data augmentation and self-distillation, can elevate Cross entropy Dice loss to surpass most topology loss functions, and that those simple methods can enhance topology loss functions as well. clDice and Skeleton Recall loss, both skeletonization-based loss functions, were also the fastest to train, making this type of loss function a promising research direction. TopoMortar and our code can be found at https://github.com/jmlipman/TopoMortar
A General Purpose Spectral Foundational Model for Both Proximal and Remote Sensing Spectral Imaging
Mar 03, 2025



Abstract:Spectral imaging data acquired via multispectral and hyperspectral cameras can have hundreds of channels, where each channel records the reflectance at a specific wavelength and bandwidth. Time and resource constraints limit our ability to collect large spectral datasets, making it difficult to build and train predictive models from scratch. In the RGB domain, we can often alleviate some of the limitations of smaller datasets by using pretrained foundational models as a starting point. However, most existing foundation models are pretrained on large datasets of 3-channel RGB images, severely limiting their effectiveness when used with spectral imaging data. The few spectral foundation models that do exist usually have one of two limitations: (1) they are built and trained only on remote sensing data limiting their application in proximal spectral imaging, (2) they utilize the more widely available multispectral imaging datasets with less than 15 channels restricting their use with hundred-channel hyperspectral images. To alleviate these issues, we propose a large-scale foundational model and dataset built upon the masked autoencoder architecture that takes advantage of spectral channel encoding, spatial-spectral masking and ImageNet pretraining for an adaptable and robust model for downstream spectral imaging tasks.
MozzaVID: Mozzarella Volumetric Image Dataset
Dec 06, 2024Abstract:Influenced by the complexity of volumetric imaging, there is a shortage of established datasets useful for benchmarking volumetric deep-learning models. As a consequence, new and existing models are not easily comparable, limiting the development of architectures optimized specifically for volumetric data. To counteract this trend, we introduce MozzaVID - a large, clean, and versatile volumetric classification dataset. Our dataset contains X-ray computed tomography (CT) images of mozzarella microstructure and enables the classification of 25 cheese types and 149 cheese samples. We provide data in three different resolutions, resulting in three dataset instances containing from 591 to 37,824 images. While being general-purpose, the dataset also facilitates investigating mozzarella structure properties. The structure of food directly affects its functional properties and thus its consumption experience. Understanding food structure helps tune the production and mimicking it enables sustainable alternatives to animal-derived food products. The complex and disordered nature of food structures brings a unique challenge, where a choice of appropriate imaging method, scale, and sample size is not trivial. With this dataset we aim to address these complexities, contributing to more robust structural analysis models. The dataset can be downloaded from: https://archive.compute.dtu.dk/files/public/projects/MozzaVID/.
Two Views Are Better than One: Monocular 3D Pose Estimation with Multiview Consistency
Nov 21, 2023



Abstract:Deducing a 3D human pose from a single 2D image or 2D keypoints is inherently challenging, given the fundamental ambiguity wherein multiple 3D poses can correspond to the same 2D representation. The acquisition of 3D data, while invaluable for resolving pose ambiguity, is expensive and requires an intricate setup, often restricting its applicability to controlled lab environments. We improve performance of monocular human pose estimation models using multiview data for fine-tuning. We propose a novel loss function, multiview consistency, to enable adding additional training data with only 2D supervision. This loss enforces that the inferred 3D pose from one view aligns with the inferred 3D pose from another view under similarity transformations. Our consistency loss substantially improves performance for fine-tuning with no available 3D data. Our experiments demonstrate that two views offset by 90 degrees are enough to obtain good performance, with only marginal improvements by adding more views. Thus, we enable the acquisition of domain-specific data by capturing activities with off-the-shelf cameras, eliminating the need for elaborate calibration procedures. This research introduces new possibilities for domain adaptation in 3D pose estimation, providing a practical and cost-effective solution to customize models for specific applications. The used dataset, featuring additional views, will be made publicly available.
SportsPose -- A Dynamic 3D sports pose dataset
Apr 04, 2023



Abstract:Accurate 3D human pose estimation is essential for sports analytics, coaching, and injury prevention. However, existing datasets for monocular pose estimation do not adequately capture the challenging and dynamic nature of sports movements. In response, we introduce SportsPose, a large-scale 3D human pose dataset consisting of highly dynamic sports movements. With more than 176,000 3D poses from 24 different subjects performing 5 different sports activities, SportsPose provides a diverse and comprehensive set of 3D poses that reflect the complex and dynamic nature of sports movements. Contrary to other markerless datasets we have quantitatively evaluated the precision of SportsPose by comparing our poses with a commercial marker-based system and achieve a mean error of 34.5 mm across all evaluation sequences. This is comparable to the error reported on the commonly used 3DPW dataset. We further introduce a new metric, local movement, which describes the movement of the wrist and ankle joints in relation to the body. With this, we show that SportsPose contains more movement than the Human3.6M and 3DPW datasets in these extremum joints, indicating that our movements are more dynamic. The dataset with accompanying code can be downloaded from our website. We hope that SportsPose will allow researchers and practitioners to develop and evaluate more effective models for the analysis of sports performance and injury prevention. With its realistic and diverse dataset, SportsPose provides a valuable resource for advancing the state-of-the-art in pose estimation in sports.
BugNIST -- A New Large Scale Volumetric 3D Image Dataset for Classification and Detection
Apr 04, 2023Abstract:Progress in 3D volumetric image analysis research is limited by the lack of datasets and most advances in analysis methods for volumetric images are based on medical data. However, medical data do not necessarily resemble the characteristics of other volumetric images such as micro-CT. To promote research in 3D volumetric image analysis beyond medical data, we have created the BugNIST dataset and made it freely available. BugNIST is an extensive dataset of micro-CT scans of 12 types of bugs, such as insects and larvae. BugNIST contains 9437 volumes where 9087 are of individual bugs and 350 are mixtures of bugs and other material. The goal of BugNIST is to benchmark classification and detection methods, and we have designed the detection challenge such that detection models are trained on scans of individual bugs and tested on bug mixtures. Models capable of solving this task will be independent of the context, i.e., the surrounding material. This is a great advantage if the context is unknown or changing, as is often the case in micro-CT. Our initial baseline analysis shows that current state-of-the-art deep learning methods classify individual bugs very well, but has great difficulty with the detection challenge. Hereby, BugNIST enables research in image analysis areas that until now have missed relevant data - both classification, detection, and hopefully more.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge