Morten Rieger Hannemose
Learning to Build Shapes by Extrusion
Jan 30, 2026Abstract:We introduce Text Encoded Extrusion (TEE), a text-based representation that expresses mesh construction as sequences of face extrusions rather than polygon lists, and a method for generating 3D meshes from TEE using a large language model (LLM). By learning extrusion sequences that assemble a mesh, similar to the way artists create meshes, our approach naturally supports arbitrary output face counts and produces manifold meshes by design, in contrast to recent transformer-based models. The learnt extrusion sequences can also be applied to existing meshes - enabling editing in addition to generation. To train our model, we decompose a library of quadrilateral meshes with non-self-intersecting face loops into constituent loops, which can be viewed as their building blocks, and finetune an LLM on the steps for reassembling the meshes by performing a sequence of extrusions. We demonstrate that our representation enables reconstruction, novel shape synthesis, and the addition of new features to existing meshes.
Towards Agnostic and Holistic Universal Image Segmentation with Bit Diffusion
Jan 06, 2026Abstract:This paper introduces a diffusion-based framework for universal image segmentation, making agnostic segmentation possible without depending on mask-based frameworks and instead predicting the full segmentation in a holistic manner. We present several key adaptations to diffusion models, which are important in this discrete setting. Notably, we show that a location-aware palette with our 2D gray code ordering improves performance. Adding a final tanh activation function is crucial for discrete data. On optimizing diffusion parameters, the sigmoid loss weighting consistently outperforms alternatives, regardless of the prediction type used, and we settle on x-prediction. While our current model does not yet surpass leading mask-based architectures, it narrows the performance gap and introduces unique capabilities, such as principled ambiguity modeling, that these models lack. All models were trained from scratch, and we believe that combining our proposed improvements with large-scale pretraining or promptable conditioning could lead to competitive models.
Diffusion Based Ambiguous Image Segmentation
Apr 08, 2025Abstract:Medical image segmentation often involves inherent uncertainty due to variations in expert annotations. Capturing this uncertainty is an important goal and previous works have used various generative image models for the purpose of representing the full distribution of plausible expert ground truths. In this work, we explore the design space of diffusion models for generative segmentation, investigating the impact of noise schedules, prediction types, and loss weightings. Notably, we find that making the noise schedule harder with input scaling significantly improves performance. We conclude that x- and v-prediction outperform epsilon-prediction, likely because the diffusion process is in the discrete segmentation domain. Many loss weightings achieve similar performance as long as they give enough weight to the end of the diffusion process. We base our experiments on the LIDC-IDRI lung lesion dataset and obtain state-of-the-art (SOTA) performance. Additionally, we introduce a randomly cropped variant of the LIDC-IDRI dataset that is better suited for uncertainty in image segmentation. Our model also achieves SOTA in this harder setting.
Two Views Are Better than One: Monocular 3D Pose Estimation with Multiview Consistency
Nov 21, 2023



Abstract:Deducing a 3D human pose from a single 2D image or 2D keypoints is inherently challenging, given the fundamental ambiguity wherein multiple 3D poses can correspond to the same 2D representation. The acquisition of 3D data, while invaluable for resolving pose ambiguity, is expensive and requires an intricate setup, often restricting its applicability to controlled lab environments. We improve performance of monocular human pose estimation models using multiview data for fine-tuning. We propose a novel loss function, multiview consistency, to enable adding additional training data with only 2D supervision. This loss enforces that the inferred 3D pose from one view aligns with the inferred 3D pose from another view under similarity transformations. Our consistency loss substantially improves performance for fine-tuning with no available 3D data. Our experiments demonstrate that two views offset by 90 degrees are enough to obtain good performance, with only marginal improvements by adding more views. Thus, we enable the acquisition of domain-specific data by capturing activities with off-the-shelf cameras, eliminating the need for elaborate calibration procedures. This research introduces new possibilities for domain adaptation in 3D pose estimation, providing a practical and cost-effective solution to customize models for specific applications. The used dataset, featuring additional views, will be made publicly available.
SportsPose -- A Dynamic 3D sports pose dataset
Apr 04, 2023



Abstract:Accurate 3D human pose estimation is essential for sports analytics, coaching, and injury prevention. However, existing datasets for monocular pose estimation do not adequately capture the challenging and dynamic nature of sports movements. In response, we introduce SportsPose, a large-scale 3D human pose dataset consisting of highly dynamic sports movements. With more than 176,000 3D poses from 24 different subjects performing 5 different sports activities, SportsPose provides a diverse and comprehensive set of 3D poses that reflect the complex and dynamic nature of sports movements. Contrary to other markerless datasets we have quantitatively evaluated the precision of SportsPose by comparing our poses with a commercial marker-based system and achieve a mean error of 34.5 mm across all evaluation sequences. This is comparable to the error reported on the commonly used 3DPW dataset. We further introduce a new metric, local movement, which describes the movement of the wrist and ankle joints in relation to the body. With this, we show that SportsPose contains more movement than the Human3.6M and 3DPW datasets in these extremum joints, indicating that our movements are more dynamic. The dataset with accompanying code can be downloaded from our website. We hope that SportsPose will allow researchers and practitioners to develop and evaluate more effective models for the analysis of sports performance and injury prevention. With its realistic and diverse dataset, SportsPose provides a valuable resource for advancing the state-of-the-art in pose estimation in sports.
Was that so hard? Estimating human classification difficulty
Mar 22, 2022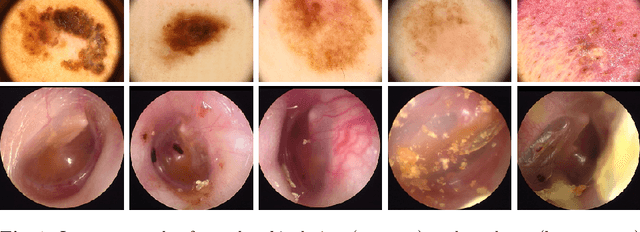
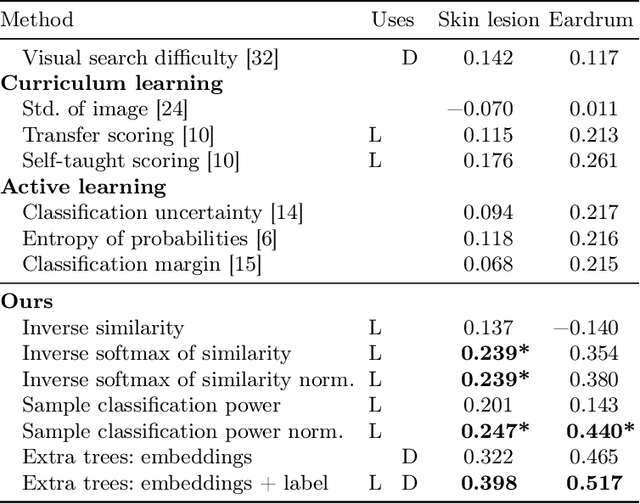

Abstract:When doctors are trained to diagnose a specific disease, they learn faster when presented with cases in order of increasing difficulty. This creates the need for automatically estimating how difficult it is for doctors to classify a given case. In this paper, we introduce methods for estimating how hard it is for a doctor to diagnose a case represented by a medical image, both when ground truth difficulties are available for training, and when they are not. Our methods are based on embeddings obtained with deep metric learning. Additionally, we introduce a practical method for obtaining ground truth human difficulty for each image case in a dataset using self-assessed certainty. We apply our methods to two different medical datasets, achieving high Kendall rank correlation coefficients, showing that we outperform existing methods by a large margin on our problem and data.
Multi-modal data generation with a deep metric variational autoencoder
Feb 07, 2022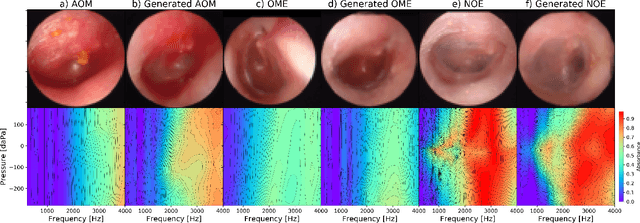
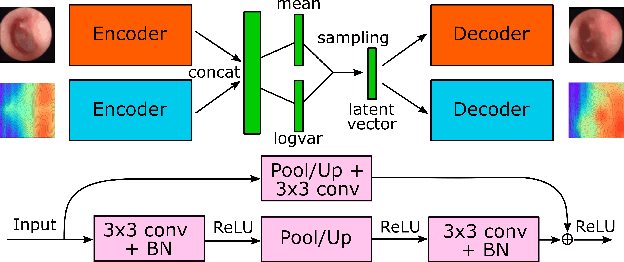
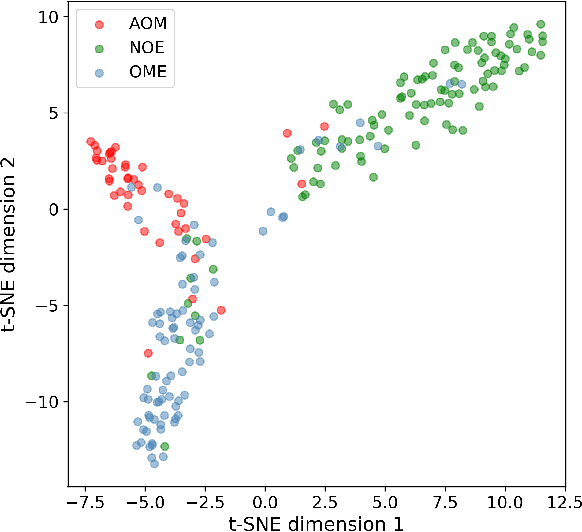
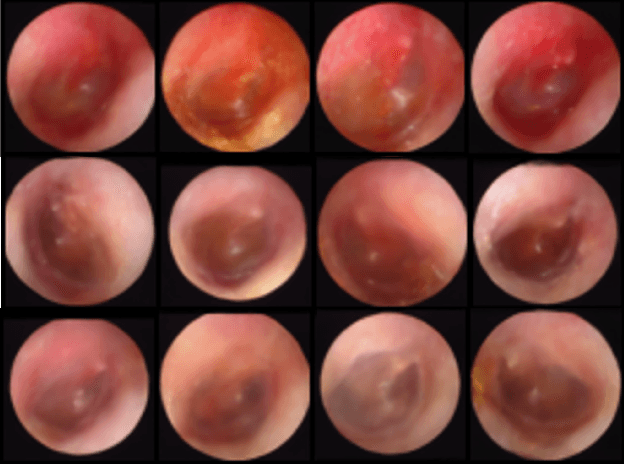
Abstract:We present a deep metric variational autoencoder for multi-modal data generation. The variational autoencoder employs triplet loss in the latent space, which allows for conditional data generation by sampling in the latent space within each class cluster. The approach is evaluated on a multi-modal dataset consisting of otoscopy images of the tympanic membrane with corresponding wideband tympanometry measurements. The modalities in this dataset are correlated, as they represent different aspects of the state of the middle ear, but they do not present a direct pixel-to-pixel correlation. The approach shows promising results for the conditional generation of pairs of images and tympanograms, and will allow for efficient data augmentation of data from multi-modal sources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge