Rasmus R. Paulsen
Geometric Deep Learning for the Assessment of Thrombosis Risk in the Left Atrial Appendage
Oct 19, 2022Abstract:The assessment of left atrial appendage (LAA) thrombogenesis has experienced major advances with the adoption of patient-specific computational fluid dynamics (CFD) simulations. Nonetheless, due to the vast computational resources and long execution times required by fluid dynamics solvers, there is an ever-growing body of work aiming to develop surrogate models of fluid flow simulations based on neural networks. The present study builds on this foundation by developing a deep learning (DL) framework capable of predicting the endothelial cell activation potential (ECAP), linked to the risk of thrombosis, solely from the patient-specific LAA geometry. To this end, we leveraged recent advancements in Geometric DL, which seamlessly extend the unparalleled potential of convolutional neural networks (CNN), to non-Euclidean data such as meshes. The model was trained with a dataset combining 202 synthetic and 54 real LAA, predicting the ECAP distributions instantaneously, with an average mean absolute error of 0.563. Moreover, the resulting framework manages to predict the anatomical features related to higher ECAP values even when trained exclusively on synthetic cases.
Was that so hard? Estimating human classification difficulty
Mar 22, 2022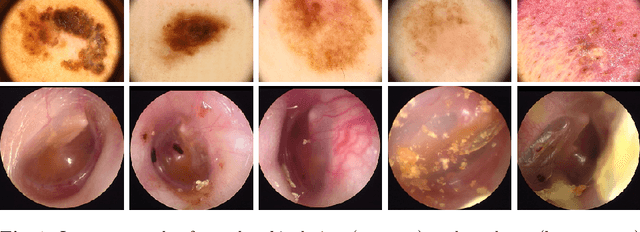
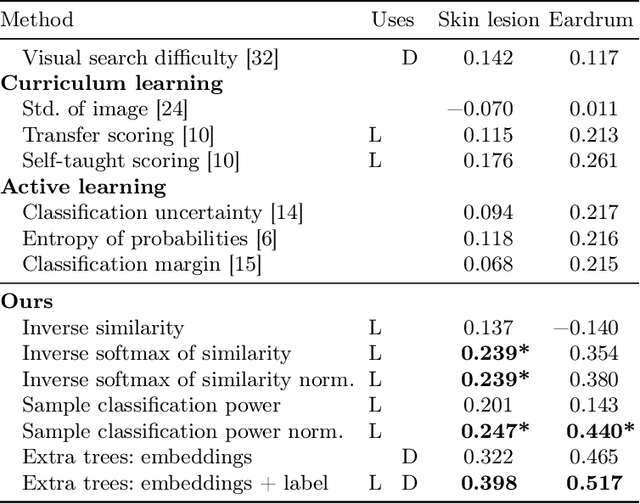

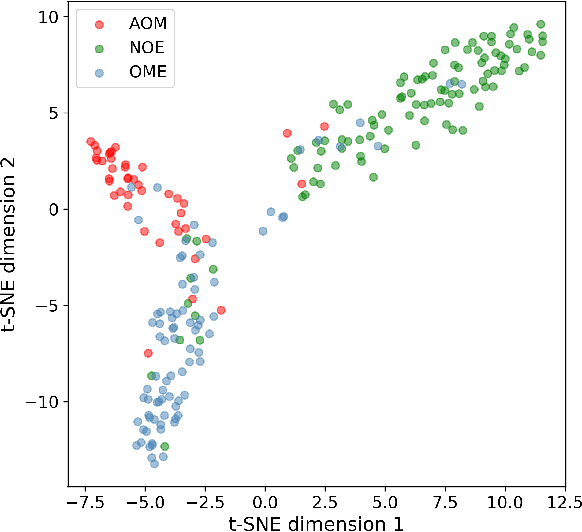
Abstract:When doctors are trained to diagnose a specific disease, they learn faster when presented with cases in order of increasing difficulty. This creates the need for automatically estimating how difficult it is for doctors to classify a given case. In this paper, we introduce methods for estimating how hard it is for a doctor to diagnose a case represented by a medical image, both when ground truth difficulties are available for training, and when they are not. Our methods are based on embeddings obtained with deep metric learning. Additionally, we introduce a practical method for obtaining ground truth human difficulty for each image case in a dataset using self-assessed certainty. We apply our methods to two different medical datasets, achieving high Kendall rank correlation coefficients, showing that we outperform existing methods by a large margin on our problem and data.
Multi-modal data generation with a deep metric variational autoencoder
Feb 07, 2022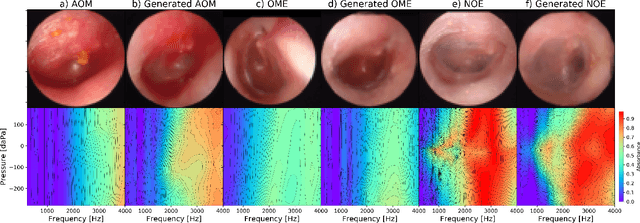
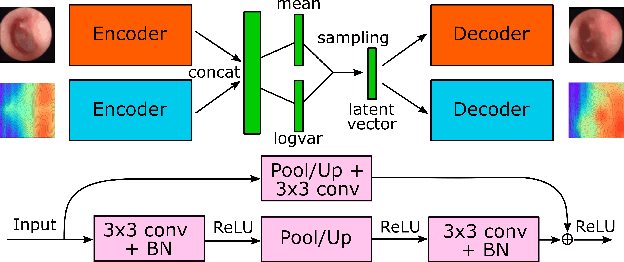
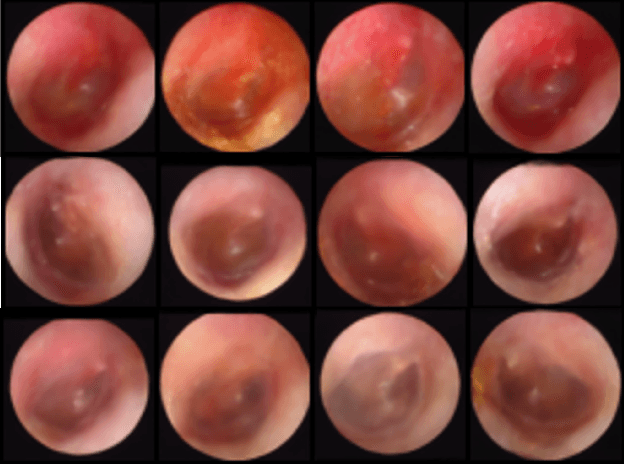
Abstract:We present a deep metric variational autoencoder for multi-modal data generation. The variational autoencoder employs triplet loss in the latent space, which allows for conditional data generation by sampling in the latent space within each class cluster. The approach is evaluated on a multi-modal dataset consisting of otoscopy images of the tympanic membrane with corresponding wideband tympanometry measurements. The modalities in this dataset are correlated, as they represent different aspects of the state of the middle ear, but they do not present a direct pixel-to-pixel correlation. The approach shows promising results for the conditional generation of pairs of images and tympanograms, and will allow for efficient data augmentation of data from multi-modal sources.
Multi-view consensus CNN for 3D facial landmark placement
Oct 14, 2019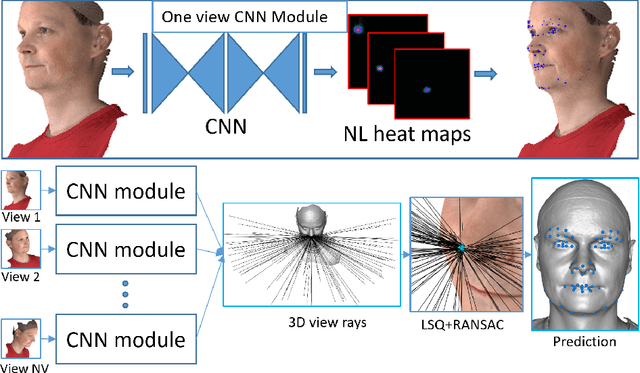
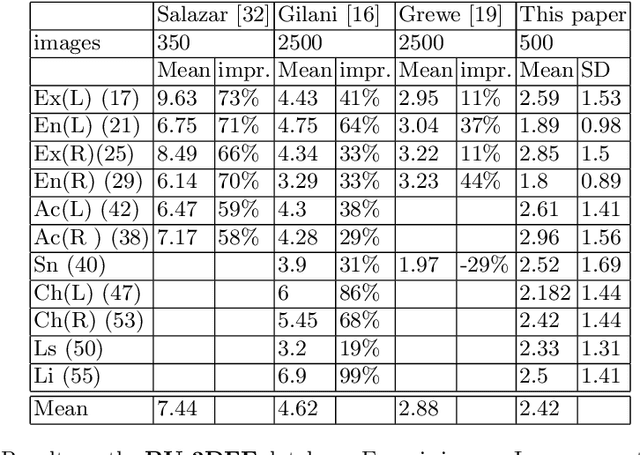
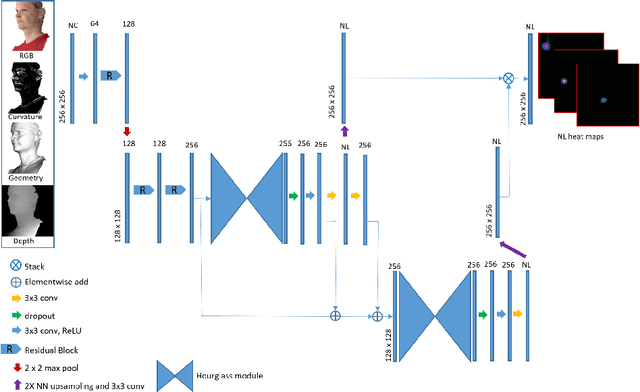
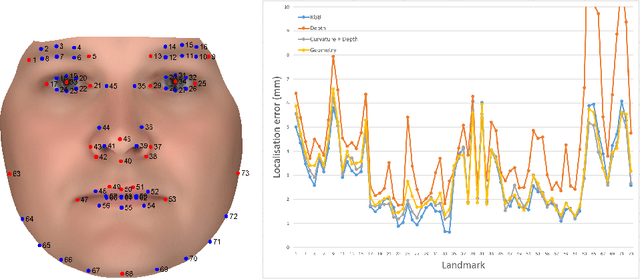
Abstract:The rapid increase in the availability of accurate 3D scanning devices has moved facial recognition and analysis into the 3D domain. 3D facial landmarks are often used as a simple measure of anatomy and it is crucial to have accurate algorithms for automatic landmark placement. The current state-of-the-art approaches have yet to gain from the dramatic increase in performance reported in human pose tracking and 2D facial landmark placement due to the use of deep convolutional neural networks (CNN). Development of deep learning approaches for 3D meshes has given rise to the new subfield called geometric deep learning, where one topic is the adaptation of meshes for the use of deep CNNs. In this work, we demonstrate how methods derived from geometric deep learning, namely multi-view CNNs, can be combined with recent advances in human pose tracking. The method finds 2D landmark estimates and propagates this information to 3D space, where a consensus method determines the accurate 3D face landmark position. We utilise the method on a standard 3D face dataset and show that it outperforms current methods by a large margin. Further, we demonstrate how models trained on 3D range scans can be used to accurately place anatomical landmarks in magnetic resonance images.
* This is a pre-print of an article published in proceedings of the asian conference on computer vision 2018 (LNCS 11361). The final authenticated version is available online at: https://doi.org/10.1007/978-3-030-20887-5_44
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge