Oscar Camara
On Safer Reinforcement Learning Policies for Sedation and Analgesia in Intensive Care
Jan 30, 2026Abstract:Pain management in intensive care usually involves complex trade-offs between therapeutic goals and patient safety, since both inadequate and excessive treatment may induce serious sequelae. Reinforcement learning can help address this challenge by learning medication dosing policies from retrospective data. However, prior work on sedation and analgesia has optimized for objectives that do not value patient survival while relying on algorithms unsuitable for imperfect information settings. We investigated the risks of these design choices by implementing a deep reinforcement learning framework to suggest hourly medication doses under partial observability. Using data from 47,144 ICU stays in the MIMIC-IV database, we trained policies to prescribe opioids, propofol, benzodiazepines, and dexmedetomidine according to two goals: reduce pain or jointly reduce pain and mortality. We found that, although the two policies were associated with lower pain, actions from the first policy were positively correlated with mortality, while those proposed by the second policy were negatively correlated. This suggests that valuing long-term outcomes could be critical for safer treatment policies, even if a short-term goal remains the primary objective.
Fetpype: An Open-Source Pipeline for Reproducible Fetal Brain MRI Analysis
Dec 19, 2025Abstract:Fetal brain Magnetic Resonance Imaging (MRI) is crucial for assessing neurodevelopment in utero. However, analyzing this data presents significant challenges due to fetal motion, low signal-to-noise ratio, and the need for complex multi-step processing, including motion correction, super-resolution reconstruction, segmentation, and surface extraction. While various specialized tools exist for individual steps, integrating them into robust, reproducible, and user-friendly workflows that go from raw images to processed volumes is not straightforward. This lack of standardization hinders reproducibility across studies and limits the adoption of advanced analysis techniques for researchers and clinicians. To address these challenges, we introduce Fetpype, an open-source Python library designed to streamline and standardize the preprocessing and analysis of T2-weighted fetal brain MRI data. Fetpype is publicly available on GitHub at https://github.com/fetpype/fetpype.
A Computational Pipeline for Advanced Analysis of 4D Flow MRI in the Left Atrium
May 14, 2025Abstract:The left atrium (LA) plays a pivotal role in modulating left ventricular filling, but our comprehension of its hemodynamics is significantly limited by the constraints of conventional ultrasound analysis. 4D flow magnetic resonance imaging (4D Flow MRI) holds promise for enhancing our understanding of atrial hemodynamics. However, the low velocities within the LA and the limited spatial resolution of 4D Flow MRI make analyzing this chamber challenging. Furthermore, the absence of dedicated computational frameworks, combined with diverse acquisition protocols and vendors, complicates gathering large cohorts for studying the prognostic value of hemodynamic parameters provided by 4D Flow MRI. In this study, we introduce the first open-source computational framework tailored for the analysis of 4D Flow MRI in the LA, enabling comprehensive qualitative and quantitative analysis of advanced hemodynamic parameters. Our framework proves robust to data from different centers of varying quality, producing high-accuracy automated segmentations (Dice $>$ 0.9 and Hausdorff 95 $<$ 3 mm), even with limited training data. Additionally, we conducted the first comprehensive assessment of energy, vorticity, and pressure parameters in the LA across a spectrum of disorders to investigate their potential as prognostic biomarkers.
NUDF: Neural Unsigned Distance Fields for high resolution 3D medical image segmentation
Apr 25, 2025Abstract:Medical image segmentation is often considered as the task of labelling each pixel or voxel as being inside or outside a given anatomy. Processing the images at their original size and resolution often result in insuperable memory requirements, but downsampling the images leads to a loss of important details. Instead of aiming to represent a smooth and continuous surface in a binary voxel-grid, we propose to learn a Neural Unsigned Distance Field (NUDF) directly from the image. The small memory requirements of NUDF allow for high resolution processing, while the continuous nature of the distance field allows us to create high resolution 3D mesh models of shapes of any topology (i.e. open surfaces). We evaluate our method on the task of left atrial appendage (LAA) segmentation from Computed Tomography (CT) images. The LAA is a complex and highly variable shape, being thus difficult to represent with traditional segmentation methods using discrete labelmaps. With our proposed method, we are able to predict 3D mesh models that capture the details of the LAA and achieve accuracy in the order of the voxel spacing in the CT images.
Spatio-temporal neural distance fields for conditional generative modeling of the heart
Jul 15, 2024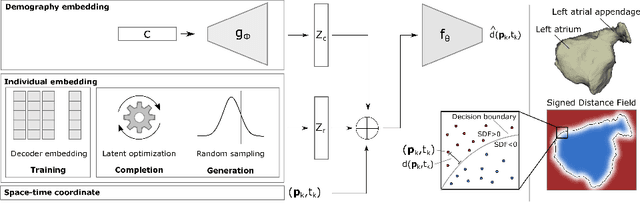
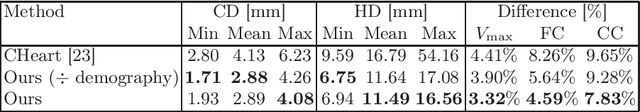
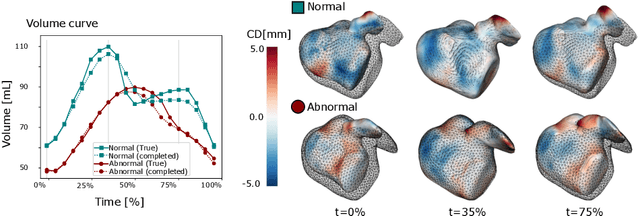
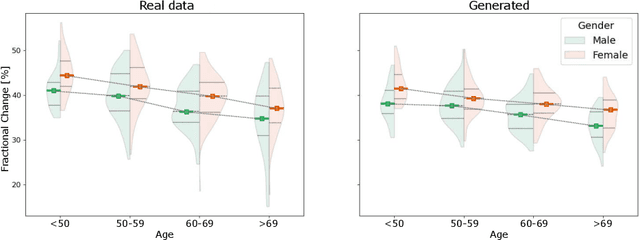
Abstract:The rhythmic pumping motion of the heart stands as a cornerstone in life, as it circulates blood to the entire human body through a series of carefully timed contractions of the individual chambers. Changes in the size, shape and movement of the chambers can be important markers for cardiac disease and modeling this in relation to clinical demography or disease is therefore of interest. Existing methods for spatio-temporal modeling of the human heart require shape correspondence over time or suffer from large memory requirements, making it difficult to use for complex anatomies. We introduce a novel conditional generative model, where the shape and movement is modeled implicitly in the form of a spatio-temporal neural distance field and conditioned on clinical demography. The model is based on an auto-decoder architecture and aims to disentangle the individual variations from that related to the clinical demography. It is tested on the left atrium (including the left atrial appendage), where it outperforms current state-of-the-art methods for anatomical sequence completion and generates synthetic sequences that realistically mimics the shape and motion of the real left atrium. In practice, this means we can infer functional measurements from a static image, generate synthetic populations with specified demography or disease and investigate how non-imaging clinical data effect the shape and motion of cardiac anatomies.
Signed Distance Field based Segmentation and Statistical Shape Modelling of the Left Atrial Appendage
Feb 12, 2024Abstract:Patients with atrial fibrillation have a 5-7 fold increased risk of having an ischemic stroke. In these cases, the most common site of thrombus localization is inside the left atrial appendage (LAA) and studies have shown a correlation between the LAA shape and the risk of ischemic stroke. These studies make use of manual measurement and qualitative assessment of shape and are therefore prone to large inter-observer discrepancies, which may explain the contradictions between the conclusions in different studies. We argue that quantitative shape descriptors are necessary to robustly characterize LAA morphology and relate to other functional parameters and stroke risk. Deep Learning methods are becoming standardly available for segmenting cardiovascular structures from high resolution images such as computed tomography (CT), but only few have been tested for LAA segmentation. Furthermore, the majority of segmentation algorithms produces non-smooth 3D models that are not ideal for further processing, such as statistical shape analysis or computational fluid modelling. In this paper we present a fully automatic pipeline for image segmentation, mesh model creation and statistical shape modelling of the LAA. The LAA anatomy is implicitly represented as a signed distance field (SDF), which is directly regressed from the CT image using Deep Learning. The SDF is further used for registering the LAA shapes to a common template and build a statistical shape model (SSM). Based on 106 automatically segmented LAAs, the built SSM reveals that the LAA shape can be quantified using approximately 5 PCA modes and allows the identification of two distinct shape clusters corresponding to the so-called chicken-wing and non-chicken-wing morphologies.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024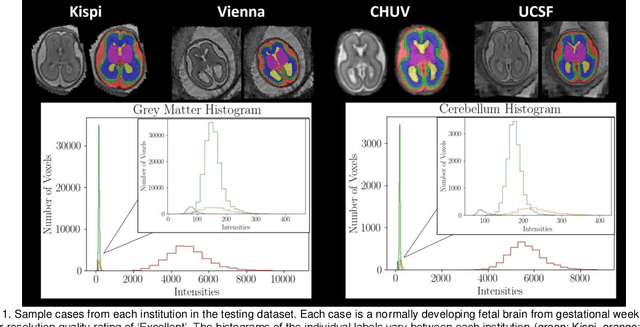
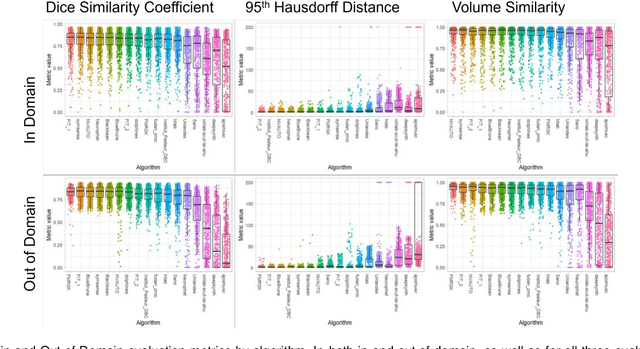
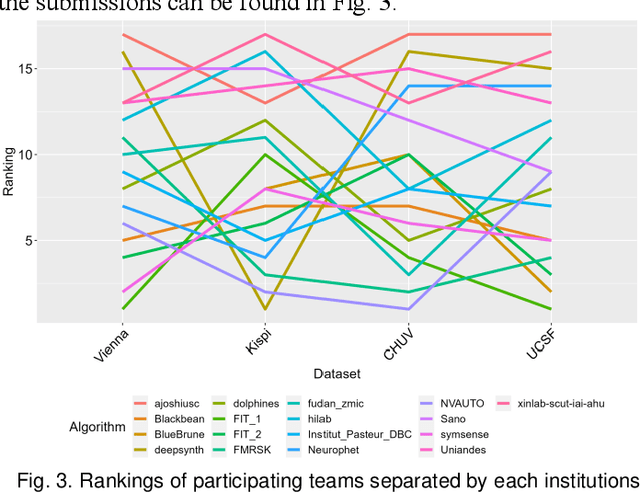
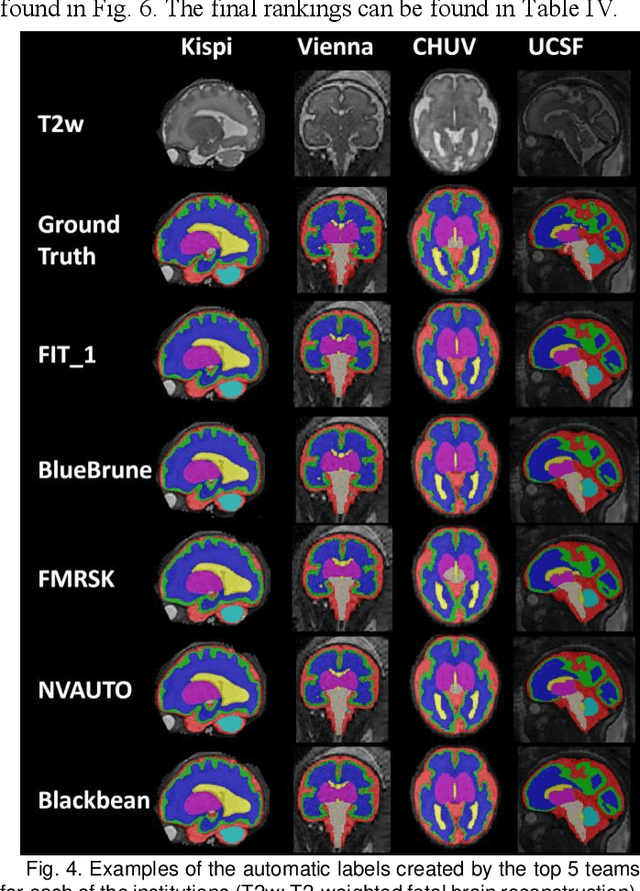
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
Unsupervised Segmentation of Fetal Brain MRI using Deep Learning Cascaded Registration
Jul 07, 2023



Abstract:Accurate segmentation of fetal brain magnetic resonance images is crucial for analyzing fetal brain development and detecting potential neurodevelopmental abnormalities. Traditional deep learning-based automatic segmentation, although effective, requires extensive training data with ground-truth labels, typically produced by clinicians through a time-consuming annotation process. To overcome this challenge, we propose a novel unsupervised segmentation method based on multi-atlas segmentation, that accurately segments multiple tissues without relying on labeled data for training. Our method employs a cascaded deep learning network for 3D image registration, which computes small, incremental deformations to the moving image to align it precisely with the fixed image. This cascaded network can then be used to register multiple annotated images with the image to be segmented, and combine the propagated labels to form a refined segmentation. Our experiments demonstrate that the proposed cascaded architecture outperforms the state-of-the-art registration methods that were tested. Furthermore, the derived segmentation method achieves similar performance and inference time to nnU-Net while only using a small subset of annotated data for the multi-atlas segmentation task and none for training the network. Our pipeline for registration and multi-atlas segmentation is publicly available at https://github.com/ValBcn/CasReg.
Geometric Deep Learning for the Assessment of Thrombosis Risk in the Left Atrial Appendage
Oct 19, 2022Abstract:The assessment of left atrial appendage (LAA) thrombogenesis has experienced major advances with the adoption of patient-specific computational fluid dynamics (CFD) simulations. Nonetheless, due to the vast computational resources and long execution times required by fluid dynamics solvers, there is an ever-growing body of work aiming to develop surrogate models of fluid flow simulations based on neural networks. The present study builds on this foundation by developing a deep learning (DL) framework capable of predicting the endothelial cell activation potential (ECAP), linked to the risk of thrombosis, solely from the patient-specific LAA geometry. To this end, we leveraged recent advancements in Geometric DL, which seamlessly extend the unparalleled potential of convolutional neural networks (CNN), to non-Euclidean data such as meshes. The model was trained with a dataset combining 202 synthetic and 54 real LAA, predicting the ECAP distributions instantaneously, with an average mean absolute error of 0.563. Moreover, the resulting framework manages to predict the anatomical features related to higher ECAP values even when trained exclusively on synthetic cases.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022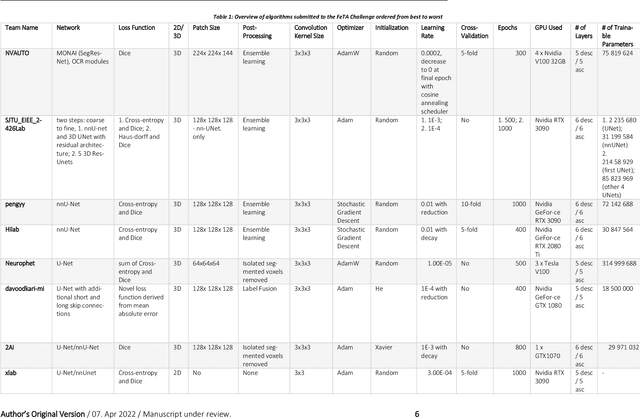
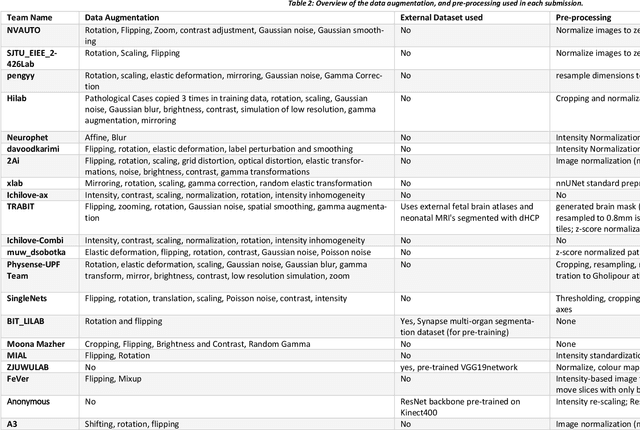
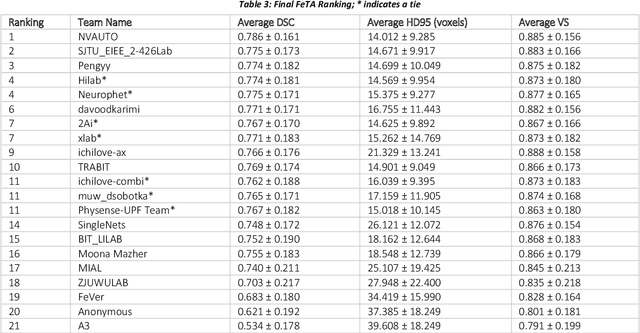
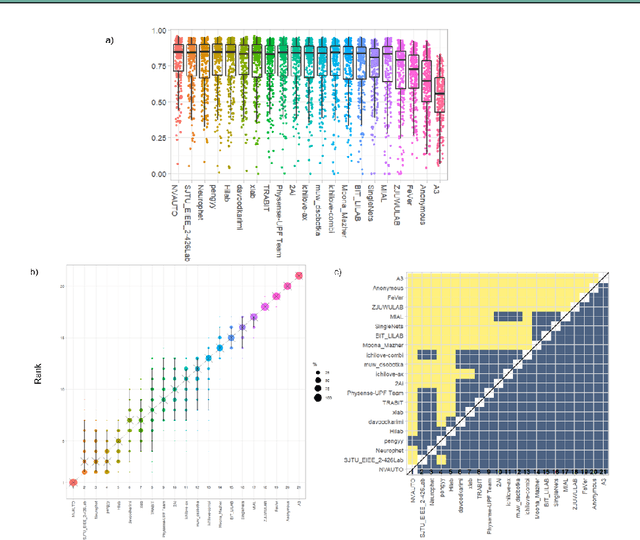
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge