Juan Acosta
Generalizing electrocardiogram delineation: training convolutional neural networks with synthetic data augmentation
Nov 25, 2021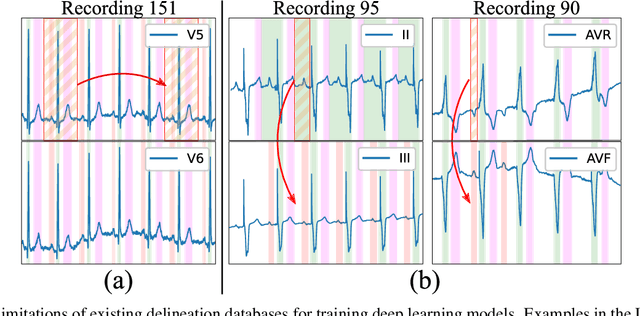
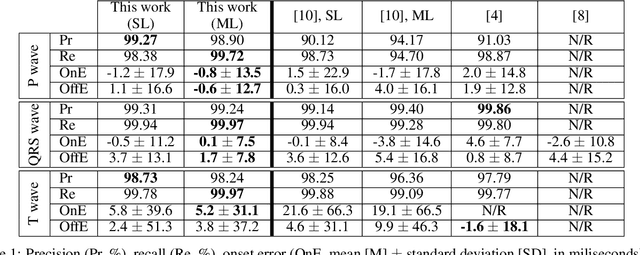
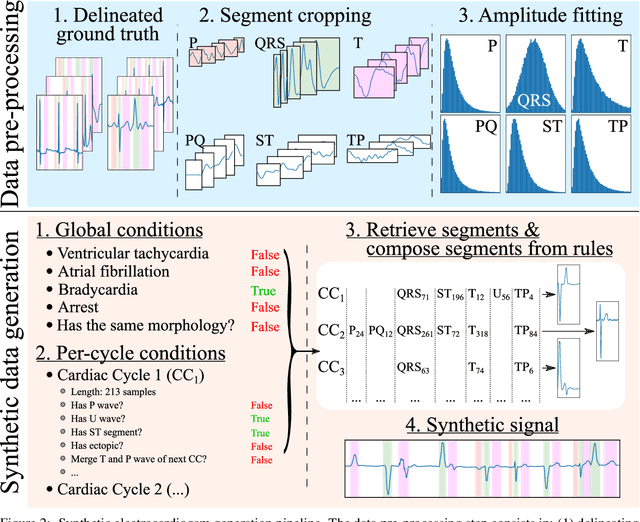
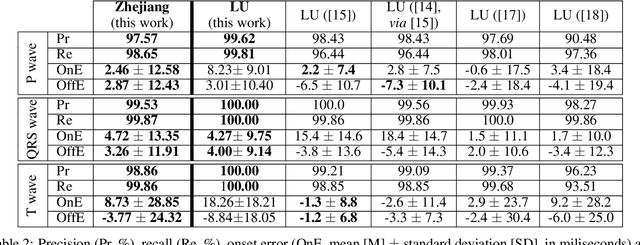
Abstract:Obtaining per-beat information is a key task in the analysis of cardiac electrocardiograms (ECG), as many downstream diagnosis tasks are dependent on ECG-based measurements. Those measurements, however, are costly to produce, especially in recordings that change throughout long periods of time. However, existing annotated databases for ECG delineation are small, being insufficient in size and in the array of pathological conditions they represent. This article delves has two main contributions. First, a pseudo-synthetic data generation algorithm was developed, based in probabilistically composing ECG traces given "pools" of fundamental segments, as cropped from the original databases, and a set of rules for their arrangement into coherent synthetic traces. The generation of conditions is controlled by imposing expert knowledge on the generated trace, which increases the input variability for training the model. Second, two novel segmentation-based loss functions have been developed, which attempt at enforcing the prediction of an exact number of independent structures and at producing closer segmentation boundaries by focusing on a reduced number of samples. The best performing model obtained an $F_1$-score of 99.38\% and a delineation error of $2.19 \pm 17.73$ ms and $4.45 \pm 18.32$ ms for all wave's fiducials (onsets and offsets, respectively), as averaged across the P, QRS and T waves for three distinct freely available databases. The excellent results were obtained despite the heterogeneous characteristics of the tested databases, in terms of lead configurations (Holter, 12-lead), sampling frequencies ($250$, $500$ and $2,000$ Hz) and represented pathophysiologies (e.g., different types of arrhythmias, sinus rhythm with structural heart disease), hinting at its generalization capabilities, while outperforming current state-of-the-art delineation approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge