Guillermo Jimenez-Perez
DiNO-Diffusion. Scaling Medical Diffusion via Self-Supervised Pre-Training
Jul 16, 2024



Abstract:Diffusion models (DMs) have emerged as powerful foundation models for a variety of tasks, with a large focus in synthetic image generation. However, their requirement of large annotated datasets for training limits their applicability in medical imaging, where datasets are typically smaller and sparsely annotated. We introduce DiNO-Diffusion, a self-supervised method for training latent diffusion models (LDMs) that conditions the generation process on image embeddings extracted from DiNO. By eliminating the reliance on annotations, our training leverages over 868k unlabelled images from public chest X-Ray (CXR) datasets. Despite being self-supervised, DiNO-Diffusion shows comprehensive manifold coverage, with FID scores as low as 4.7, and emerging properties when evaluated in downstream tasks. It can be used to generate semantically-diverse synthetic datasets even from small data pools, demonstrating up to 20% AUC increase in classification performance when used for data augmentation. Images were generated with different sampling strategies over the DiNO embedding manifold and using real images as a starting point. Results suggest, DiNO-Diffusion could facilitate the creation of large datasets for flexible training of downstream AI models from limited amount of real data, while also holding potential for privacy preservation. Additionally, DiNO-Diffusion demonstrates zero-shot segmentation performance of up to 84.4% Dice score when evaluating lung lobe segmentation. This evidences good CXR image-anatomy alignment, akin to segmenting using textual descriptors on vanilla DMs. Finally, DiNO-Diffusion can be easily adapted to other medical imaging modalities or state-of-the-art diffusion models, opening the door for large-scale, multi-domain image generation pipelines for medical imaging.
Latent Diffusion Models with Image-Derived Annotations for Enhanced AI-Assisted Cancer Diagnosis in Histopathology
Dec 15, 2023Abstract:Artificial Intelligence (AI) based image analysis has an immense potential to support diagnostic histopathology, including cancer diagnostics. However, developing supervised AI methods requires large-scale annotated datasets. A potentially powerful solution is to augment training data with synthetic data. Latent diffusion models, which can generate high-quality, diverse synthetic images, are promising. However, the most common implementations rely on detailed textual descriptions, which are not generally available in this domain. This work proposes a method that constructs structured textual prompts from automatically extracted image features. We experiment with the PCam dataset, composed of tissue patches only loosely annotated as healthy or cancerous. We show that including image-derived features in the prompt, as opposed to only healthy and cancerous labels, improves the Fr\'echet Inception Distance (FID) from 178.8 to 90.2. We also show that pathologists find it challenging to detect synthetic images, with a median sensitivity/specificity of 0.55/0.55. Finally, we show that synthetic data effectively trains AI models.
Generalizing electrocardiogram delineation: training convolutional neural networks with synthetic data augmentation
Nov 25, 2021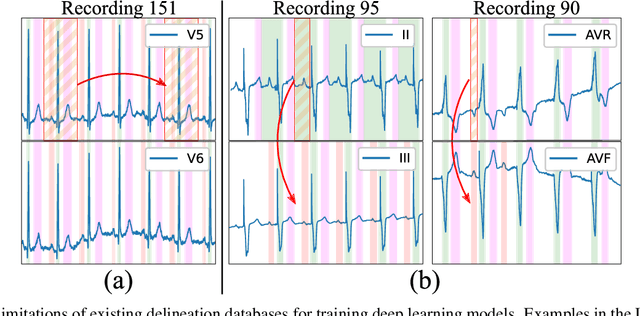
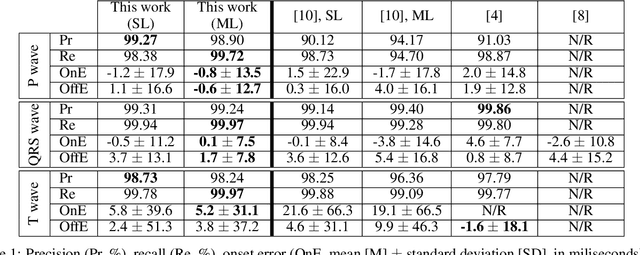
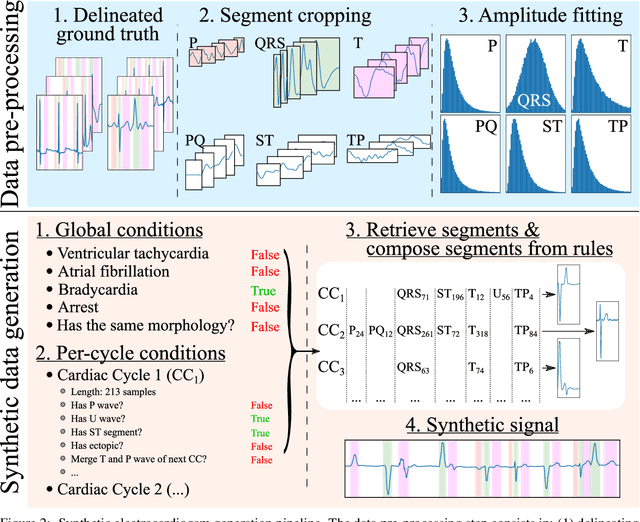
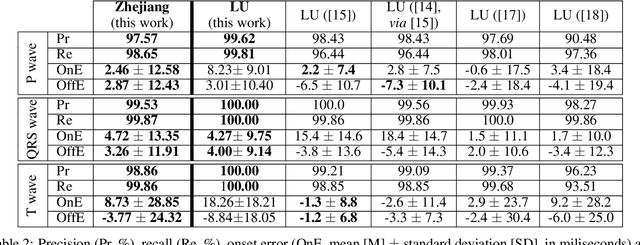
Abstract:Obtaining per-beat information is a key task in the analysis of cardiac electrocardiograms (ECG), as many downstream diagnosis tasks are dependent on ECG-based measurements. Those measurements, however, are costly to produce, especially in recordings that change throughout long periods of time. However, existing annotated databases for ECG delineation are small, being insufficient in size and in the array of pathological conditions they represent. This article delves has two main contributions. First, a pseudo-synthetic data generation algorithm was developed, based in probabilistically composing ECG traces given "pools" of fundamental segments, as cropped from the original databases, and a set of rules for their arrangement into coherent synthetic traces. The generation of conditions is controlled by imposing expert knowledge on the generated trace, which increases the input variability for training the model. Second, two novel segmentation-based loss functions have been developed, which attempt at enforcing the prediction of an exact number of independent structures and at producing closer segmentation boundaries by focusing on a reduced number of samples. The best performing model obtained an $F_1$-score of 99.38\% and a delineation error of $2.19 \pm 17.73$ ms and $4.45 \pm 18.32$ ms for all wave's fiducials (onsets and offsets, respectively), as averaged across the P, QRS and T waves for three distinct freely available databases. The excellent results were obtained despite the heterogeneous characteristics of the tested databases, in terms of lead configurations (Holter, 12-lead), sampling frequencies ($250$, $500$ and $2,000$ Hz) and represented pathophysiologies (e.g., different types of arrhythmias, sinus rhythm with structural heart disease), hinting at its generalization capabilities, while outperforming current state-of-the-art delineation approaches.
ECG-DelNet: Delineation of Ambulatory Electrocardiograms with Mixed Quality Labeling Using Neural Networks
May 11, 2020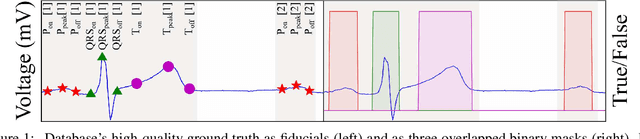
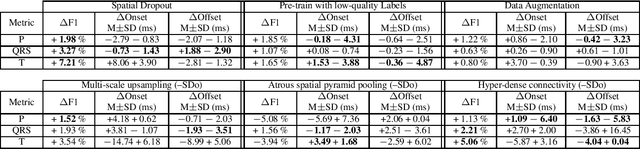
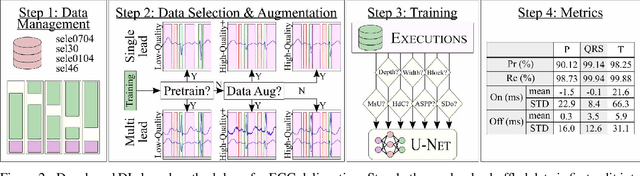
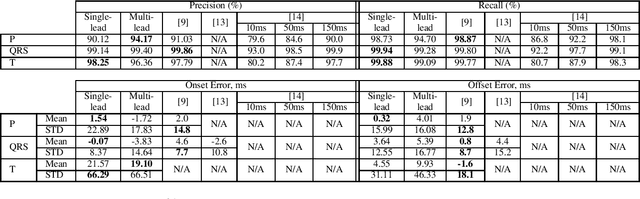
Abstract:Electrocardiogram (ECG) detection and delineation are key steps for numerous tasks in clinical practice, as ECG is the most performed non-invasive test for assessing cardiac condition. State-of-the-art algorithms employ digital signal processing (DSP), which require laborious rule adaptation to new morphologies. In contrast, deep learning (DL) algorithms, especially for classification, are gaining weight in academic and industrial settings. However, the lack of model explainability and small databases hinder their applicability. We demonstrate DL can be successfully applied to low interpretative tasks by embedding ECG detection and delineation onto a segmentation framework. For this purpose, we adapted and validated the most used neural network architecture for image segmentation, the U-Net, to one-dimensional data. The model was trained using PhysioNet's QT database, comprised of 105 ambulatory ECG recordings, for single- and multi-lead scenarios. To alleviate data scarcity, data regularization techniques such as pre-training with low-quality data labels, performing ECG-based data augmentation and applying strong model regularizers to the architecture were attempted. Other variations in the model's capacity (U-Net's depth and width), alongside the application of state-of-the-art additions, were evaluated. These variations were exhaustively validated in a 5-fold cross-validation manner. The best performing configuration reached precisions of 90.12%, 99.14% and 98.25% and recalls of 98.73%, 99.94% and 99.88% for the P, QRS and T waves, respectively, on par with DSP-based approaches. Despite being a data-hungry technique trained on a small dataset, DL-based approaches demonstrate to be a viable alternative to traditional DSP-based ECG processing techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge