Sadegh Mohammadi
Investigating the Effects of Diffusion-based Conditional Generative Speech Models Used for Speech Enhancement on Dysarthric Speech
Dec 18, 2024Abstract:In this study, we aim to explore the effect of pre-trained conditional generative speech models for the first time on dysarthric speech due to Parkinson's disease recorded in an ideal/non-noisy condition. Considering one category of generative models, i.e., diffusion-based speech enhancement, these models are previously trained to learn the distribution of clean (i.e, recorded in a noise-free environment) typical speech signals. Therefore, we hypothesized that when being exposed to dysarthric speech they might remove the unseen atypical paralinguistic cues during the enhancement process. By considering the automatic dysarthric speech detection task, in this study, we experimentally show that during the enhancement process of dysarthric speech data recorded in an ideal non-noisy environment, some of the acoustic dysarthric speech cues are lost. Therefore such pre-trained models are not yet suitable in the context of dysarthric speech enhancement since they manipulate the pathological speech cues when they process clean dysarthric speech. Furthermore, we show that the removed acoustics cues by the enhancement models in the form of residue speech signal can provide complementary dysarthric cues when fused with the original input speech signal in the feature space.
Panoptic Segmentation of Mammograms with Text-To-Image Diffusion Model
Jul 19, 2024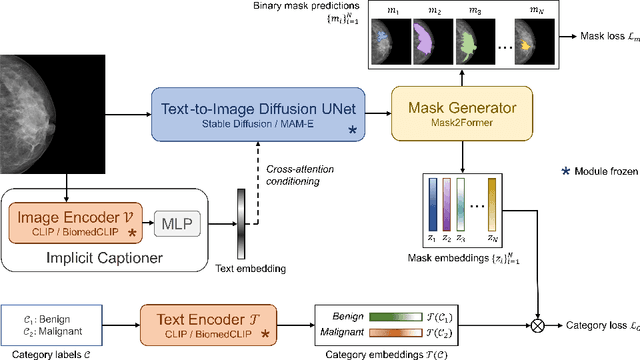
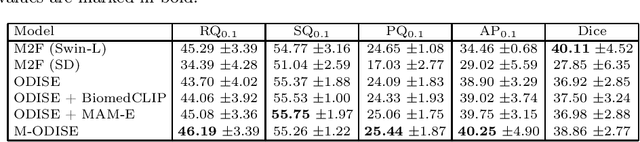
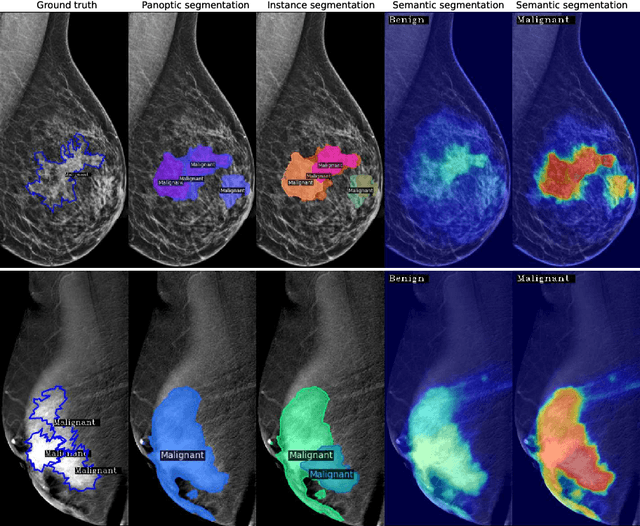
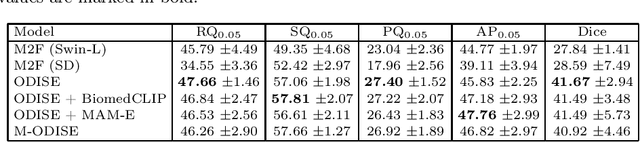
Abstract:Mammography is crucial for breast cancer surveillance and early diagnosis. However, analyzing mammography images is a demanding task for radiologists, who often review hundreds of mammograms daily, leading to overdiagnosis and overtreatment. Computer-Aided Diagnosis (CAD) systems have been developed to assist in this process, but their capabilities, particularly in lesion segmentation, remained limited. With the contemporary advances in deep learning their performance may be improved. Recently, vision-language diffusion models emerged, demonstrating outstanding performance in image generation and transferability to various downstream tasks. We aim to harness their capabilities for breast lesion segmentation in a panoptic setting, which encompasses both semantic and instance-level predictions. Specifically, we propose leveraging pretrained features from a Stable Diffusion model as inputs to a state-of-the-art panoptic segmentation architecture, resulting in accurate delineation of individual breast lesions. To bridge the gap between natural and medical imaging domains, we incorporated a mammography-specific MAM-E diffusion model and BiomedCLIP image and text encoders into this framework. We evaluated our approach on two recently published mammography datasets, CDD-CESM and VinDr-Mammo. For the instance segmentation task, we noted 40.25 AP0.1 and 46.82 AP0.05, as well as 25.44 PQ0.1 and 26.92 PQ0.05. For the semantic segmentation task, we achieved Dice scores of 38.86 and 40.92, respectively.
DiNO-Diffusion. Scaling Medical Diffusion via Self-Supervised Pre-Training
Jul 16, 2024



Abstract:Diffusion models (DMs) have emerged as powerful foundation models for a variety of tasks, with a large focus in synthetic image generation. However, their requirement of large annotated datasets for training limits their applicability in medical imaging, where datasets are typically smaller and sparsely annotated. We introduce DiNO-Diffusion, a self-supervised method for training latent diffusion models (LDMs) that conditions the generation process on image embeddings extracted from DiNO. By eliminating the reliance on annotations, our training leverages over 868k unlabelled images from public chest X-Ray (CXR) datasets. Despite being self-supervised, DiNO-Diffusion shows comprehensive manifold coverage, with FID scores as low as 4.7, and emerging properties when evaluated in downstream tasks. It can be used to generate semantically-diverse synthetic datasets even from small data pools, demonstrating up to 20% AUC increase in classification performance when used for data augmentation. Images were generated with different sampling strategies over the DiNO embedding manifold and using real images as a starting point. Results suggest, DiNO-Diffusion could facilitate the creation of large datasets for flexible training of downstream AI models from limited amount of real data, while also holding potential for privacy preservation. Additionally, DiNO-Diffusion demonstrates zero-shot segmentation performance of up to 84.4% Dice score when evaluating lung lobe segmentation. This evidences good CXR image-anatomy alignment, akin to segmenting using textual descriptors on vanilla DMs. Finally, DiNO-Diffusion can be easily adapted to other medical imaging modalities or state-of-the-art diffusion models, opening the door for large-scale, multi-domain image generation pipelines for medical imaging.
Latent Diffusion Models with Image-Derived Annotations for Enhanced AI-Assisted Cancer Diagnosis in Histopathology
Dec 15, 2023Abstract:Artificial Intelligence (AI) based image analysis has an immense potential to support diagnostic histopathology, including cancer diagnostics. However, developing supervised AI methods requires large-scale annotated datasets. A potentially powerful solution is to augment training data with synthetic data. Latent diffusion models, which can generate high-quality, diverse synthetic images, are promising. However, the most common implementations rely on detailed textual descriptions, which are not generally available in this domain. This work proposes a method that constructs structured textual prompts from automatically extracted image features. We experiment with the PCam dataset, composed of tissue patches only loosely annotated as healthy or cancerous. We show that including image-derived features in the prompt, as opposed to only healthy and cancerous labels, improves the Fr\'echet Inception Distance (FID) from 178.8 to 90.2. We also show that pathologists find it challenging to detect synthetic images, with a median sensitivity/specificity of 0.55/0.55. Finally, we show that synthetic data effectively trains AI models.
Towards Hierarchical Regional Transformer-based Multiple Instance Learning
Aug 24, 2023



Abstract:The classification of gigapixel histopathology images with deep multiple instance learning models has become a critical task in digital pathology and precision medicine. In this work, we propose a Transformer-based multiple instance learning approach that replaces the traditional learned attention mechanism with a regional, Vision Transformer inspired self-attention mechanism. We present a method that fuses regional patch information to derive slide-level predictions and show how this regional aggregation can be stacked to hierarchically process features on different distance levels. To increase predictive accuracy, especially for datasets with small, local morphological features, we introduce a method to focus the image processing on high attention regions during inference. Our approach is able to significantly improve performance over the baseline on two histopathology datasets and points towards promising directions for further research.
FAIR4Cov: Fused Audio Instance and Representation for COVID-19 Detection
Apr 22, 2022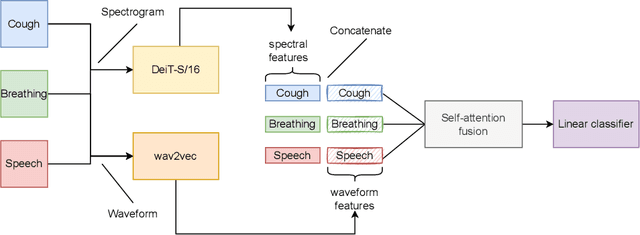



Abstract:Audio-based classification techniques on body sounds have long been studied to support diagnostic decisions, particularly in pulmonary diseases. In response to the urgency of the COVID-19 pandemic, a growing number of models are developed to identify COVID-19 patients based on acoustic input. Most models focus on cough because the dry cough is the best-known symptom of COVID-19. However, other body sounds, such as breath and speech, have also been revealed to correlate with COVID-19 as well. In this work, rather than relying on a specific body sound, we propose Fused Audio Instance and Representation for COVID-19 Detection (FAIR4Cov). It relies on constructing a joint feature vector obtained from a plurality of body sounds in waveform and spectrogram representation. The core component of FAIR4Cov is a self-attention fusion unit that is trained to establish the relation of multiple body sounds and audio representations and integrate it into a compact feature vector. We set up our experiments on different combinations of body sounds using only waveform, spectrogram, and a joint representation of waveform and spectrogram. Our findings show that the use of self-attention to combine extracted features from cough, breath, and speech sounds leads to the best performance with an Area Under the Receiver Operating Characteristic Curve (AUC) score of 0.8658, a sensitivity of 0.8057, and a specificity of 0.7958. This AUC is 0.0227 higher than the one of the models trained on spectrograms only and 0.0847 higher than the one of the models trained on waveforms only. The results demonstrate that the combination of spectrogram with waveform representation helps to enrich the extracted features and outperforms the models with single representation.
How Transferable Are Self-supervised Features in Medical Image Classification Tasks?
Aug 23, 2021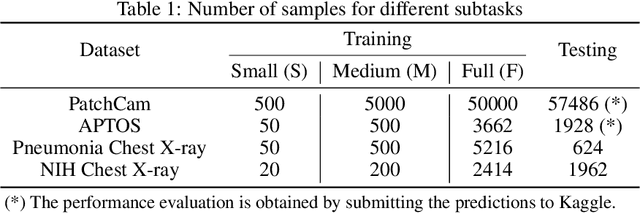


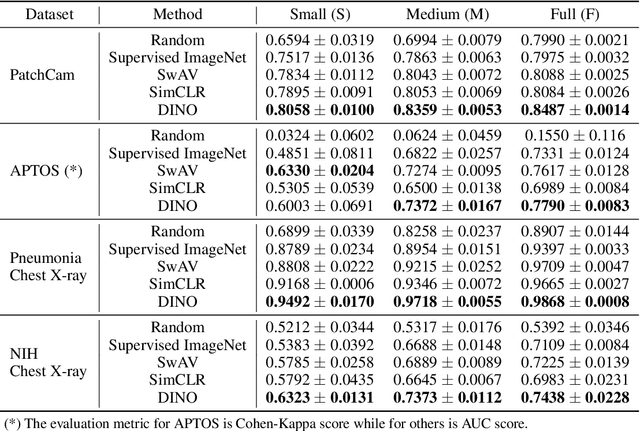
Abstract:Transfer learning has become a standard practice to mitigate the lack of labeled data in medical classification tasks. Whereas finetuning a downstream task using supervised ImageNet pretrained features is straightforward and extensively investigated in many works, there is little study on the usefulness of self-supervised pretraining. In this paper, we assess the transferability of ImageNet self-supervisedpretraining by evaluating the performance of models initialized with pretrained features from three self-supervised techniques (SimCLR, SwAV, and DINO) on selected medical classification tasks. The chosen tasks cover tumor detection in sentinel axillary lymph node images, diabetic retinopathy classification in fundus images, and multiple pathological condition classification in chest X-ray images. We demonstrate that self-supervised pretrained models yield richer embeddings than their supervised counterpart, which benefits downstream tasks in view of both linear evaluation and finetuning. For example, in view of linear evaluation at acritically small subset of the data, we see an improvement up to 14.79% in Kappa score in the diabetic retinopathy classification task, 5.4% in AUC in the tumor classification task, 7.03% AUC in the pneumonia detection, and 9.4% in AUC in the detection of pathological conditions in chest X-ray. In addition, we introduce Dynamic Visual Meta-Embedding (DVME) as an end-to-end transfer learning approach that fuses pretrained embeddings from multiple models. We show that the collective representation obtained by DVME leads to a significant improvement in the performance of selected tasks compared to using a single pretrained model approach and can be generalized to any combination of pretrained models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge