Matthias Lenga
Zero-shot System for Automatic Body Region Detection for Volumetric CT and MR Images
Feb 09, 2026Abstract:Reliable identification of anatomical body regions is a prerequisite for many automated medical imaging workflows, yet existing solutions remain heavily dependent on unreliable DICOM metadata. Current solutions mainly use supervised learning, which limits their applicability in many real-world scenarios. In this work, we investigate whether body region detection in volumetric CT and MR images can be achieved in a fully zero-shot manner by using knowledge embedded in large pre-trained foundation models. We propose and systematically evaluate three training-free pipelines: (1) a segmentation-driven rule-based system leveraging pre-trained multi-organ segmentation models, (2) a Multimodal Large Language Model (MLLM) guided by radiologist-defined rules, and (3) a segmentation-aware MLLM that combines visual input with explicit anatomical evidence. All methods are evaluated on 887 heterogeneous CT and MR scans with manually verified anatomical region labels. The segmentation-driven rule-based approach achieves the strongest and most consistent performance, with weighted F1-scores of 0.947 (CT) and 0.914 (MR), demonstrating robustness across modalities and atypical scan coverage. The MLLM performs competitively in visually distinctive regions, while the segmentation-aware MLLM reveals fundamental limitations.
Content-based 3D Image Retrieval and a ColBERT-inspired Re-ranking for Tumor Flagging and Staging
Jul 23, 2025Abstract:The increasing volume of medical images poses challenges for radiologists in retrieving relevant cases. Content-based image retrieval (CBIR) systems offer potential for efficient access to similar cases, yet lack standardized evaluation and comprehensive studies. Building on prior studies for tumor characterization via CBIR, this study advances CBIR research for volumetric medical images through three key contributions: (1) a framework eliminating reliance on pre-segmented data and organ-specific datasets, aligning with large and unstructured image archiving systems, i.e. PACS in clinical practice; (2) introduction of C-MIR, a novel volumetric re-ranking method adapting ColBERT's contextualized late interaction mechanism for 3D medical imaging; (3) comprehensive evaluation across four tumor sites using three feature extractors and three database configurations. Our evaluations highlight the significant advantages of C-MIR. We demonstrate the successful adaptation of the late interaction principle to volumetric medical images, enabling effective context-aware re-ranking. A key finding is C-MIR's ability to effectively localize the region of interest, eliminating the need for pre-segmentation of datasets and offering a computationally efficient alternative to systems relying on expensive data enrichment steps. C-MIR demonstrates promising improvements in tumor flagging, achieving improved performance, particularly for colon and lung tumors (p<0.05). C-MIR also shows potential for improving tumor staging, warranting further exploration of its capabilities. Ultimately, our work seeks to bridge the gap between advanced retrieval techniques and their practical applications in healthcare, paving the way for improved diagnostic processes.
Exploring AI-based System Design for Pixel-level Protected Health Information Detection in Medical Images
Jan 16, 2025Abstract:De-identification of medical images is a critical step to ensure privacy during data sharing in research and clinical settings. The initial step in this process involves detecting Protected Health Information (PHI), which can be found in image metadata or imprinted within image pixels. Despite the importance of such systems, there has been limited evaluation of existing AI-based solutions, creating barriers to the development of reliable and robust tools. In this study, we present an AI-based pipeline for PHI detection, comprising three key components: text detection, text extraction, and analysis of PHI content in medical images. By experimenting with exchanging roles of vision and language models within the pipeline, we evaluate the performance and recommend the best setup for the PHI detection task.
Five Pitfalls When Assessing Synthetic Medical Images with Reference Metrics
Aug 12, 2024Abstract:Reference metrics have been developed to objectively and quantitatively compare two images. Especially for evaluating the quality of reconstructed or compressed images, these metrics have shown very useful. Extensive tests of such metrics on benchmarks of artificially distorted natural images have revealed which metric best correlate with human perception of quality. Direct transfer of these metrics to the evaluation of generative models in medical imaging, however, can easily lead to pitfalls, because assumptions about image content, image data format and image interpretation are often very different. Also, the correlation of reference metrics and human perception of quality can vary strongly for different kinds of distortions and commonly used metrics, such as SSIM, PSNR and MAE are not the best choice for all situations. We selected five pitfalls that showcase unexpected and probably undesired reference metric scores and discuss strategies to avoid them.
Similarity Metrics for MR Image-To-Image Translation
May 15, 2024Abstract:Image-to-image translation can create large impact in medical imaging, i.e. if images of a patient can be translated to another modality, type or sequence for better diagnosis. However, these methods must be validated by human reader studies, which are costly and restricted to small samples. Automatic evaluation of large samples to pre-evaluate and continuously improve methods before human validation is needed. In this study, we give an overview of reference and non-reference metrics for image synthesis assessment and investigate the ability of nine metrics, that need a reference (SSIM, MS-SSIM, PSNR, MSE, NMSE, MAE, LPIPS, NMI and PCC) and three non-reference metrics (BLUR, MSN, MNG) to detect 11 kinds of distortions in MR images from the BraSyn dataset. In addition we test a downstream segmentation metric and the effect of three normalization methods (Minmax, cMinMax and Zscore). Although PSNR and SSIM are frequently used to evaluate generative models for image-to-image-translation tasks in the medical domain, they show very specific shortcomings. SSIM ignores blurring but is very sensitive to intensity shifts in unnormalized MR images. PSNR is even more sensitive to different normalization methods and hardly measures the degree of distortions. Further metrics, such as LPIPS, NMI and DICE can be very useful to evaluate other similarity aspects. If the images to be compared are misaligned, most metrics are flawed. By carefully selecting and reasonably combining image similarity metrics, the training and selection of generative models for MR image synthesis can be improved. Many aspects of their output can be validated before final and costly evaluation by trained radiologists is conducted.
Content-Based Image Retrieval for Multi-Class Volumetric Radiology Images: A Benchmark Study
May 15, 2024Abstract:While content-based image retrieval (CBIR) has been extensively studied in natural image retrieval, its application to medical images presents ongoing challenges, primarily due to the 3D nature of medical images. Recent studies have shown the potential use of pre-trained vision embeddings for CBIR in the context of radiology image retrieval. However, a benchmark for the retrieval of 3D volumetric medical images is still lacking, hindering the ability to objectively evaluate and compare the efficiency of proposed CBIR approaches in medical imaging. In this study, we extend previous work and establish a benchmark for region-based and multi-organ retrieval using the TotalSegmentator dataset (TS) with detailed multi-organ annotations. We benchmark embeddings derived from pre-trained supervised models on medical images against embeddings derived from pre-trained unsupervised models on non-medical images for 29 coarse and 104 detailed anatomical structures in volume and region levels. We adopt a late interaction re-ranking method inspired by text matching for image retrieval, comparing it against the original method proposed for volume and region retrieval achieving retrieval recall of 1.0 for diverse anatomical regions with a wide size range. The findings and methodologies presented in this paper provide essential insights and benchmarks for the development and evaluation of CBIR approaches in the context of medical imaging.
BraSyn 2023 challenge: Missing MRI synthesis and the effect of different learning objectives
Mar 18, 2024Abstract:This work addresses the Brain Magnetic Resonance Image Synthesis for Tumor Segmentation (BraSyn) challenge, which was hosted as part of the Brain Tumor Segmentation (BraTS) challenge in 2023. In this challenge, researchers are invited to synthesize a missing magnetic resonance image sequence, given other available sequences, to facilitate tumor segmentation pipelines trained on complete sets of image sequences. This problem can be tackled using deep learning within the framework of paired image-to-image translation. In this study, we propose investigating the effectiveness of a commonly used deep learning framework, such as Pix2Pix, trained under the supervision of different image-quality loss functions. Our results indicate that the use of different loss functions significantly affects the synthesis quality. We systematically study the impact of various loss functions in the multi-sequence MR image synthesis setting of the BraSyn challenge. Furthermore, we demonstrate how image synthesis performance can be optimized by combining different learning objectives beneficially.
Benchmarking Pretrained Vision Embeddings for Near- and Duplicate Detection in Medical Images
Dec 12, 2023Abstract:Near- and duplicate image detection is a critical concern in the field of medical imaging. Medical datasets often contain similar or duplicate images from various sources, which can lead to significant performance issues and evaluation biases, especially in machine learning tasks due to data leakage between training and testing subsets. In this paper, we present an approach for identifying near- and duplicate 3D medical images leveraging publicly available 2D computer vision embeddings. We assessed our approach by comparing embeddings extracted from two state-of-the-art self-supervised pretrained models and two different vector index structures for similarity retrieval. We generate an experimental benchmark based on the publicly available Medical Segmentation Decathlon dataset. The proposed method yields promising results for near- and duplicate image detection achieving a mean sensitivity and specificity of 0.9645 and 0.8559, respectively.
Medical Image Retrieval Using Pretrained Embeddings
Nov 22, 2023Abstract:A wide range of imaging techniques and data formats available for medical images make accurate retrieval from image databases challenging. Efficient retrieval systems are crucial in advancing medical research, enabling large-scale studies and innovative diagnostic tools. Thus, addressing the challenges of medical image retrieval is essential for the continued enhancement of healthcare and research. In this study, we evaluated the feasibility of employing four state-of-the-art pretrained models for medical image retrieval at modality, body region, and organ levels and compared the results of two similarity indexing approaches. Since the employed networks take 2D images, we analyzed the impacts of weighting and sampling strategies to incorporate 3D information during retrieval of 3D volumes. We showed that medical image retrieval is feasible using pretrained networks without any additional training or fine-tuning steps. Using pretrained embeddings, we achieved a recall of 1 for various tasks at modality, body region, and organ level.
Uncertainty Estimation in Contrast-Enhanced MR Image Translation with Multi-Axis Fusion
Nov 20, 2023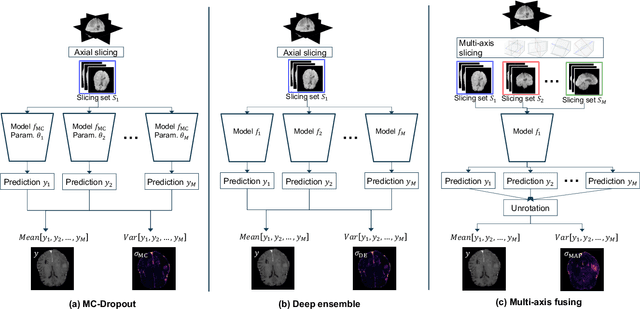
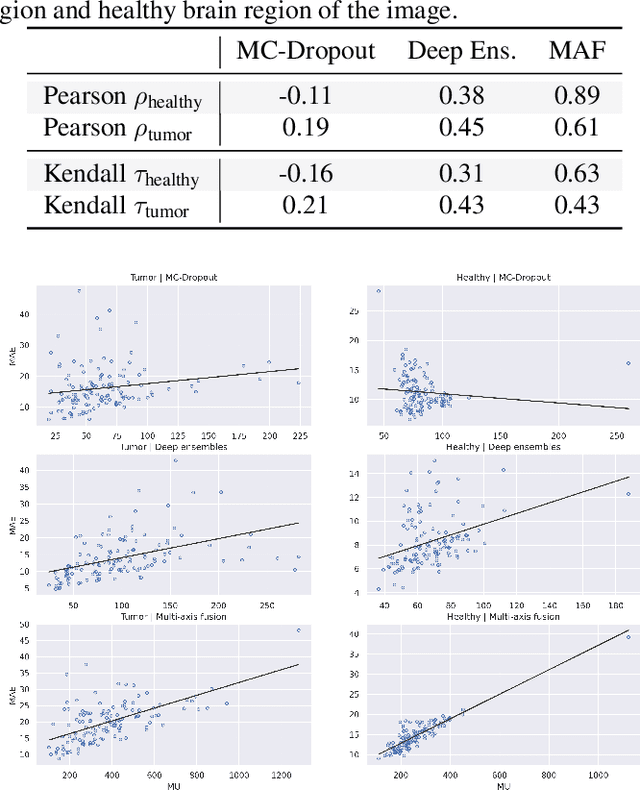
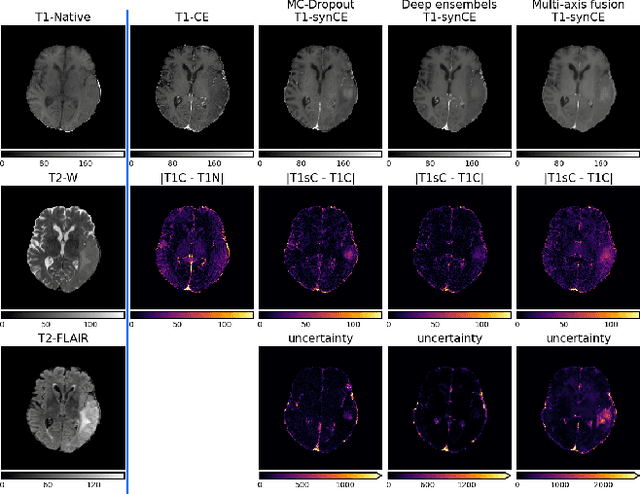
Abstract:In recent years, deep learning has been applied to a wide range of medical imaging and image processing tasks. In this work, we focus on the estimation of epistemic uncertainty for 3D medical image-to-image translation. We propose a novel model uncertainty quantification method, Multi-Axis Fusion (MAF), which relies on the integration of complementary information derived from multiple views on volumetric image data. The proposed approach is applied to the task of synthesizing contrast enhanced T1-weighted images based on native T1, T2 and T2-FLAIR scans. The quantitative findings indicate a strong correlation ($\rho_{\text healthy} = 0.89$) between the mean absolute image synthetization error and the mean uncertainty score for our MAF method. Hence, we consider MAF as a promising approach to solve the highly relevant task of detecting synthetization failures at inference time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge