Steffen Vogler
Content-based 3D Image Retrieval and a ColBERT-inspired Re-ranking for Tumor Flagging and Staging
Jul 23, 2025Abstract:The increasing volume of medical images poses challenges for radiologists in retrieving relevant cases. Content-based image retrieval (CBIR) systems offer potential for efficient access to similar cases, yet lack standardized evaluation and comprehensive studies. Building on prior studies for tumor characterization via CBIR, this study advances CBIR research for volumetric medical images through three key contributions: (1) a framework eliminating reliance on pre-segmented data and organ-specific datasets, aligning with large and unstructured image archiving systems, i.e. PACS in clinical practice; (2) introduction of C-MIR, a novel volumetric re-ranking method adapting ColBERT's contextualized late interaction mechanism for 3D medical imaging; (3) comprehensive evaluation across four tumor sites using three feature extractors and three database configurations. Our evaluations highlight the significant advantages of C-MIR. We demonstrate the successful adaptation of the late interaction principle to volumetric medical images, enabling effective context-aware re-ranking. A key finding is C-MIR's ability to effectively localize the region of interest, eliminating the need for pre-segmentation of datasets and offering a computationally efficient alternative to systems relying on expensive data enrichment steps. C-MIR demonstrates promising improvements in tumor flagging, achieving improved performance, particularly for colon and lung tumors (p<0.05). C-MIR also shows potential for improving tumor staging, warranting further exploration of its capabilities. Ultimately, our work seeks to bridge the gap between advanced retrieval techniques and their practical applications in healthcare, paving the way for improved diagnostic processes.
DiNO-Diffusion. Scaling Medical Diffusion via Self-Supervised Pre-Training
Jul 16, 2024



Abstract:Diffusion models (DMs) have emerged as powerful foundation models for a variety of tasks, with a large focus in synthetic image generation. However, their requirement of large annotated datasets for training limits their applicability in medical imaging, where datasets are typically smaller and sparsely annotated. We introduce DiNO-Diffusion, a self-supervised method for training latent diffusion models (LDMs) that conditions the generation process on image embeddings extracted from DiNO. By eliminating the reliance on annotations, our training leverages over 868k unlabelled images from public chest X-Ray (CXR) datasets. Despite being self-supervised, DiNO-Diffusion shows comprehensive manifold coverage, with FID scores as low as 4.7, and emerging properties when evaluated in downstream tasks. It can be used to generate semantically-diverse synthetic datasets even from small data pools, demonstrating up to 20% AUC increase in classification performance when used for data augmentation. Images were generated with different sampling strategies over the DiNO embedding manifold and using real images as a starting point. Results suggest, DiNO-Diffusion could facilitate the creation of large datasets for flexible training of downstream AI models from limited amount of real data, while also holding potential for privacy preservation. Additionally, DiNO-Diffusion demonstrates zero-shot segmentation performance of up to 84.4% Dice score when evaluating lung lobe segmentation. This evidences good CXR image-anatomy alignment, akin to segmenting using textual descriptors on vanilla DMs. Finally, DiNO-Diffusion can be easily adapted to other medical imaging modalities or state-of-the-art diffusion models, opening the door for large-scale, multi-domain image generation pipelines for medical imaging.
Content-Based Image Retrieval for Multi-Class Volumetric Radiology Images: A Benchmark Study
May 15, 2024Abstract:While content-based image retrieval (CBIR) has been extensively studied in natural image retrieval, its application to medical images presents ongoing challenges, primarily due to the 3D nature of medical images. Recent studies have shown the potential use of pre-trained vision embeddings for CBIR in the context of radiology image retrieval. However, a benchmark for the retrieval of 3D volumetric medical images is still lacking, hindering the ability to objectively evaluate and compare the efficiency of proposed CBIR approaches in medical imaging. In this study, we extend previous work and establish a benchmark for region-based and multi-organ retrieval using the TotalSegmentator dataset (TS) with detailed multi-organ annotations. We benchmark embeddings derived from pre-trained supervised models on medical images against embeddings derived from pre-trained unsupervised models on non-medical images for 29 coarse and 104 detailed anatomical structures in volume and region levels. We adopt a late interaction re-ranking method inspired by text matching for image retrieval, comparing it against the original method proposed for volume and region retrieval achieving retrieval recall of 1.0 for diverse anatomical regions with a wide size range. The findings and methodologies presented in this paper provide essential insights and benchmarks for the development and evaluation of CBIR approaches in the context of medical imaging.
Croissant: A Metadata Format for ML-Ready Datasets
Mar 28, 2024Abstract:Data is a critical resource for Machine Learning (ML), yet working with data remains a key friction point. This paper introduces Croissant, a metadata format for datasets that simplifies how data is used by ML tools and frameworks. Croissant makes datasets more discoverable, portable and interoperable, thereby addressing significant challenges in ML data management and responsible AI. Croissant is already supported by several popular dataset repositories, spanning hundreds of thousands of datasets, ready to be loaded into the most popular ML frameworks.
Medical Image Retrieval Using Pretrained Embeddings
Nov 22, 2023Abstract:A wide range of imaging techniques and data formats available for medical images make accurate retrieval from image databases challenging. Efficient retrieval systems are crucial in advancing medical research, enabling large-scale studies and innovative diagnostic tools. Thus, addressing the challenges of medical image retrieval is essential for the continued enhancement of healthcare and research. In this study, we evaluated the feasibility of employing four state-of-the-art pretrained models for medical image retrieval at modality, body region, and organ levels and compared the results of two similarity indexing approaches. Since the employed networks take 2D images, we analyzed the impacts of weighting and sampling strategies to incorporate 3D information during retrieval of 3D volumes. We showed that medical image retrieval is feasible using pretrained networks without any additional training or fine-tuning steps. Using pretrained embeddings, we achieved a recall of 1 for various tasks at modality, body region, and organ level.
Towards Fine-grained Visual Representations by Combining Contrastive Learning with Image Reconstruction and Attention-weighted Pooling
Apr 09, 2021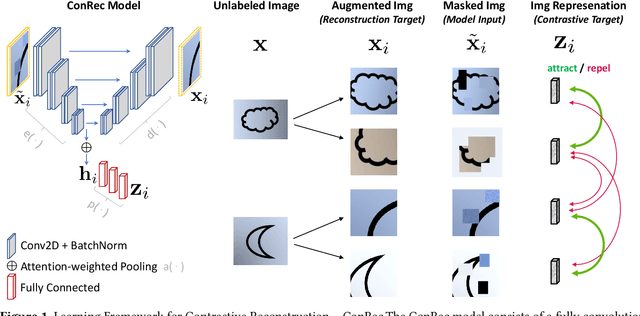
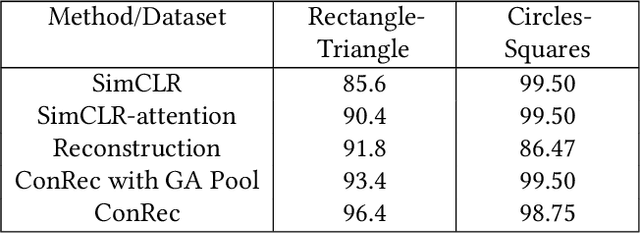
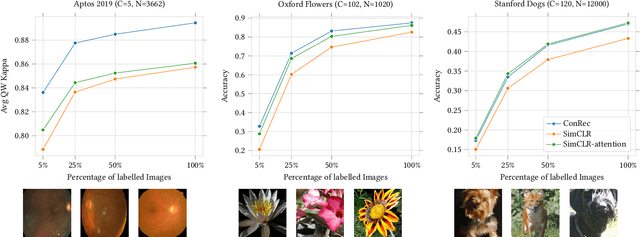
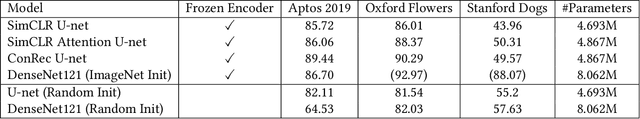
Abstract:This paper presents Contrastive Reconstruction, ConRec - a self-supervised learning algorithm that obtains image representations by jointly optimizing a contrastive and a self-reconstruction loss. We showcase that state-of-the-art contrastive learning methods (e.g. SimCLR) have shortcomings to capture fine-grained visual features in their representations. ConRec extends the SimCLR framework by adding (1) a self-reconstruction task and (2) an attention mechanism within the contrastive learning task. This is accomplished by applying a simple encoder-decoder architecture with two heads. We show that both extensions contribute towards an improved vector representation for images with fine-grained visual features. Combining those concepts, ConRec outperforms SimCLR and SimCLR with Attention-Pooling on fine-grained classification datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge