Ivo M. Baltruschat
Exploring AI-based System Design for Pixel-level Protected Health Information Detection in Medical Images
Jan 16, 2025Abstract:De-identification of medical images is a critical step to ensure privacy during data sharing in research and clinical settings. The initial step in this process involves detecting Protected Health Information (PHI), which can be found in image metadata or imprinted within image pixels. Despite the importance of such systems, there has been limited evaluation of existing AI-based solutions, creating barriers to the development of reliable and robust tools. In this study, we present an AI-based pipeline for PHI detection, comprising three key components: text detection, text extraction, and analysis of PHI content in medical images. By experimenting with exchanging roles of vision and language models within the pipeline, we evaluate the performance and recommend the best setup for the PHI detection task.
Five Pitfalls When Assessing Synthetic Medical Images with Reference Metrics
Aug 12, 2024Abstract:Reference metrics have been developed to objectively and quantitatively compare two images. Especially for evaluating the quality of reconstructed or compressed images, these metrics have shown very useful. Extensive tests of such metrics on benchmarks of artificially distorted natural images have revealed which metric best correlate with human perception of quality. Direct transfer of these metrics to the evaluation of generative models in medical imaging, however, can easily lead to pitfalls, because assumptions about image content, image data format and image interpretation are often very different. Also, the correlation of reference metrics and human perception of quality can vary strongly for different kinds of distortions and commonly used metrics, such as SSIM, PSNR and MAE are not the best choice for all situations. We selected five pitfalls that showcase unexpected and probably undesired reference metric scores and discuss strategies to avoid them.
BraSyn 2023 challenge: Missing MRI synthesis and the effect of different learning objectives
Mar 18, 2024Abstract:This work addresses the Brain Magnetic Resonance Image Synthesis for Tumor Segmentation (BraSyn) challenge, which was hosted as part of the Brain Tumor Segmentation (BraTS) challenge in 2023. In this challenge, researchers are invited to synthesize a missing magnetic resonance image sequence, given other available sequences, to facilitate tumor segmentation pipelines trained on complete sets of image sequences. This problem can be tackled using deep learning within the framework of paired image-to-image translation. In this study, we propose investigating the effectiveness of a commonly used deep learning framework, such as Pix2Pix, trained under the supervision of different image-quality loss functions. Our results indicate that the use of different loss functions significantly affects the synthesis quality. We systematically study the impact of various loss functions in the multi-sequence MR image synthesis setting of the BraSyn challenge. Furthermore, we demonstrate how image synthesis performance can be optimized by combining different learning objectives beneficially.
Uncertainty Estimation in Contrast-Enhanced MR Image Translation with Multi-Axis Fusion
Nov 20, 2023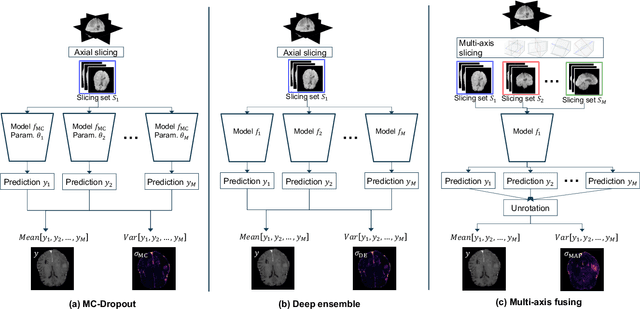
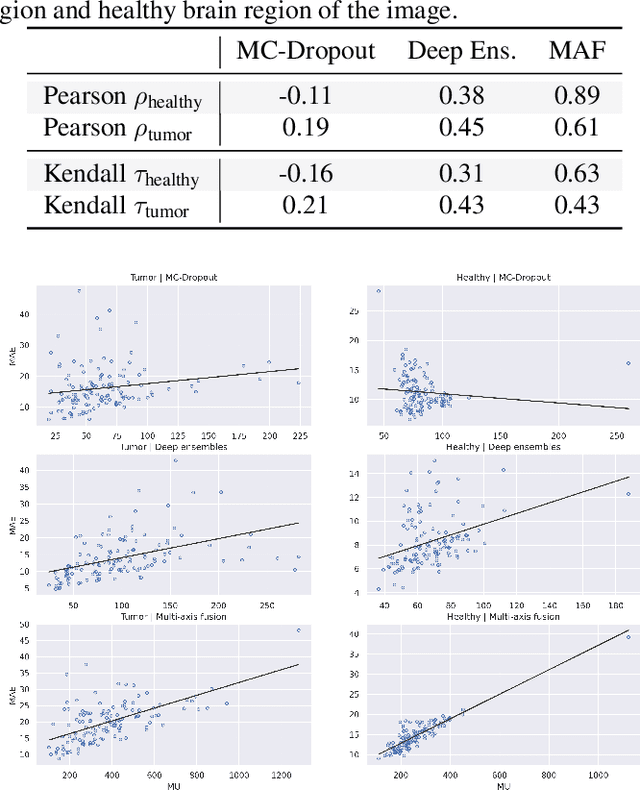
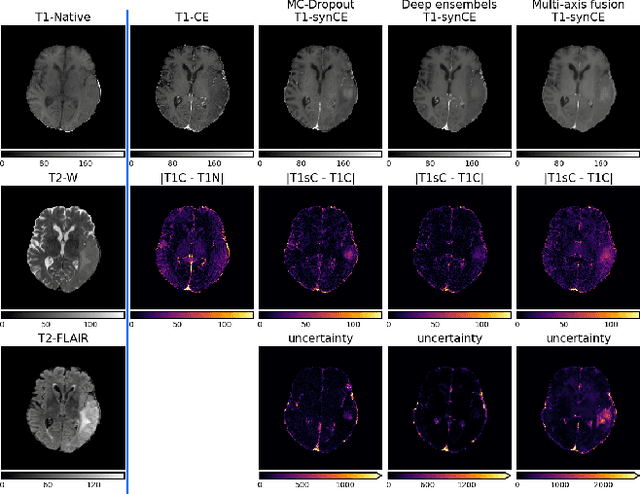
Abstract:In recent years, deep learning has been applied to a wide range of medical imaging and image processing tasks. In this work, we focus on the estimation of epistemic uncertainty for 3D medical image-to-image translation. We propose a novel model uncertainty quantification method, Multi-Axis Fusion (MAF), which relies on the integration of complementary information derived from multiple views on volumetric image data. The proposed approach is applied to the task of synthesizing contrast enhanced T1-weighted images based on native T1, T2 and T2-FLAIR scans. The quantitative findings indicate a strong correlation ($\rho_{\text healthy} = 0.89$) between the mean absolute image synthetization error and the mean uncertainty score for our MAF method. Hence, we consider MAF as a promising approach to solve the highly relevant task of detecting synthetization failures at inference time.
fRegGAN with K-space Loss Regularization for Medical Image Translation
Mar 28, 2023



Abstract:Generative adversarial networks (GANs) have shown remarkable success in generating realistic images and are increasingly used in medical imaging for image-to-image translation tasks. However, GANs tend to suffer from a frequency bias towards low frequencies, which can lead to the removal of important structures in the generated images. To address this issue, we propose a novel frequency-aware image-to-image translation framework based on the supervised RegGAN approach, which we call fRegGAN. The framework employs a K-space loss to regularize the frequency content of the generated images and incorporates well-known properties of MRI K-space geometry to guide the network training process. By combine our method with the RegGAN approach, we can mitigate the effect of training with misaligned data and frequency bias at the same time. We evaluate our method on the public BraTS dataset and outperform the baseline methods in terms of both quantitative and qualitative metrics when synthesizing T2-weighted from T1-weighted MR images. Detailed ablation studies are provided to understand the effect of each modification on the final performance. The proposed method is a step towards improving the performance of image-to-image translation and synthesis in the medical domain and shows promise for other applications in the field of image processing and generation.
Intelligent Chest X-ray Worklist Prioritization by CNNs: A Clinical Workflow Simulation
Jan 23, 2020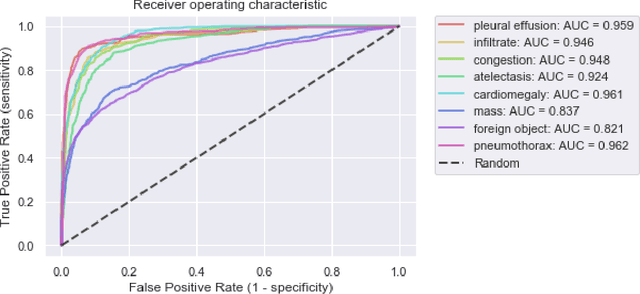
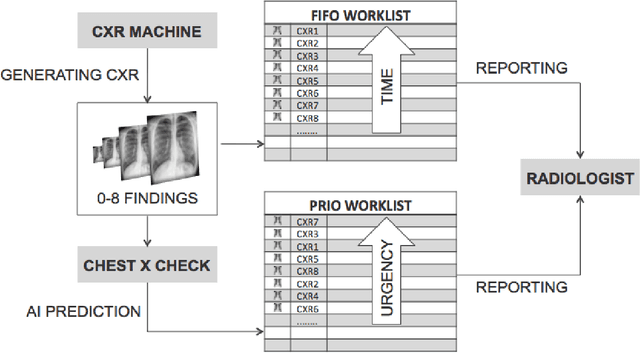
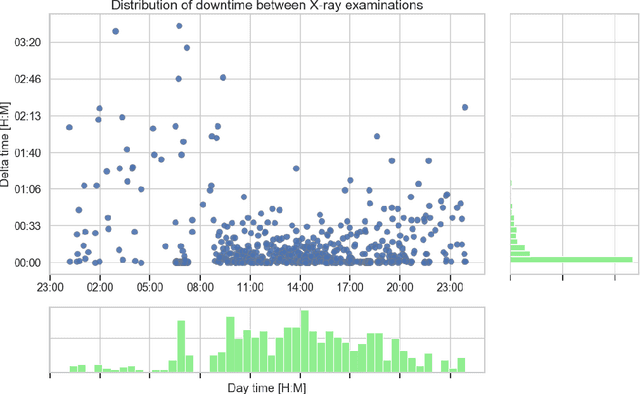
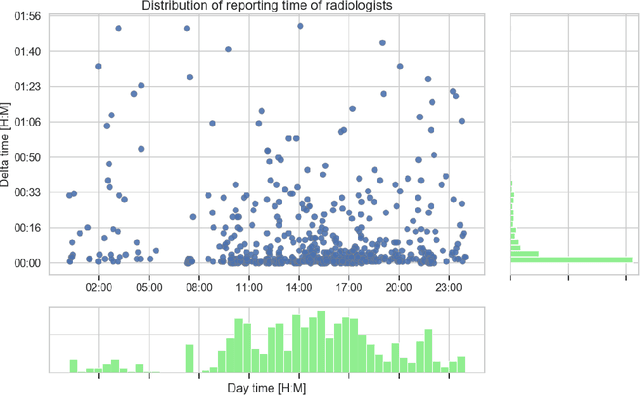
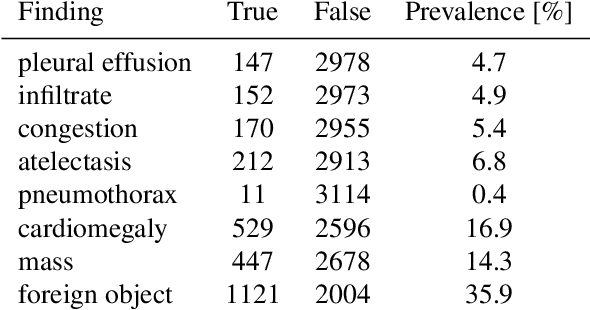
Abstract:Growing radiologic workload and shortage of medical experts worldwide often lead to delayed or even unreported examinations, which poses a risk for patient's safety in case of unrecognized findings in chest radiographs (CXR). The aim was to evaluate, whether deep learning algorithms for an intelligent worklist prioritization might optimize the radiology workflow and can reduce report turnaround times (RTAT) for critical findings, instead of reporting according to the First-In-First-Out-Principle (FIFO). Furthermore, we investigated the problem of false negative prediction in the context of worklist prioritization. To assess the potential benefit of an intelligent worklist prioritization, three different workflow simulations based on our analysis were run and RTAT were compared: FIFO (non-prioritized), Prio1 (prioritized) and Prio2 (prioritized, with RTATmax.). Examination triage was performed by "ChestXCheck", a convolutional neural network, classifying eight different pathological findings ranked in descending order of urgency: pneumothorax, pleural effusion, infiltrate, congestion, atelectasis, cardiomegaly, mass and foreign object. The average RTAT for all critical findings was significantly reduced by both Prio simulations compared to the FIFO simulation (e.g. pneumothorax: 32.1 min vs. 69.7 min; p < 0.0001), while the average RTAT for normal examinations increased at the same time (69.5 min vs. 90.0 min; p < 0.0001). Both effects were slightly lower at Prio2 than at Prio1, whereas the maximum RTAT at Prio1 was substantially higher for all classes, due to individual examinations rated false negative.Our Prio2 simulation demonstrated that intelligent worklist prioritization by deep learning algorithms reduces average RTAT for critical findings in chest X-ray while maintaining a similar maximum RTAT as FIFO.
When does Bone Suppression and Lung Field Segmentation Improve Chest X-Ray Disease Classification?
Oct 17, 2018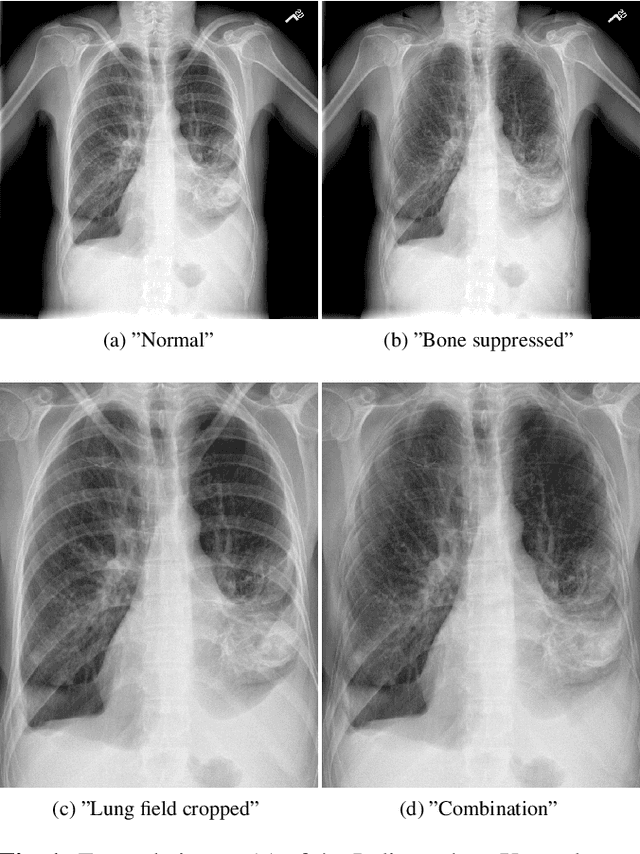
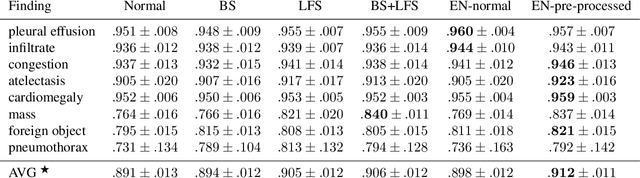
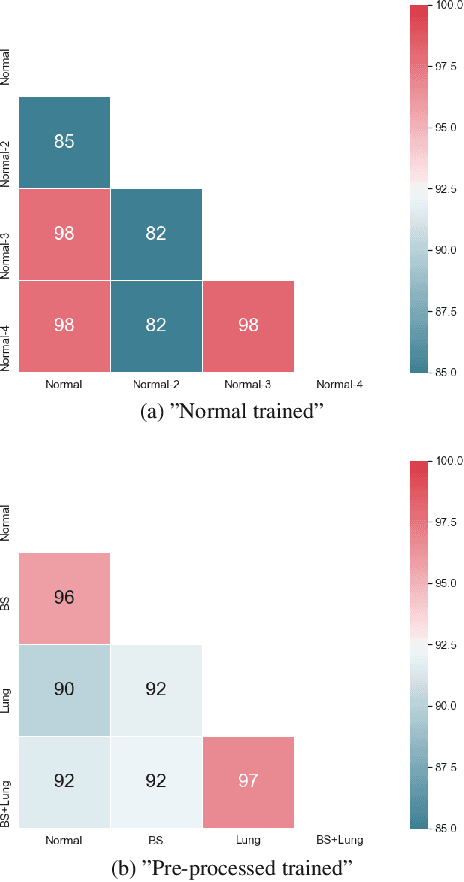
Abstract:Chest radiography is the most common clinical examination type. To improve the quality of patient care and to reduce workload, methods for automatic pathology classification have been developed. In this contribution we investigate the usefulness of two advanced image pre-processing techniques, initially developed for image reading by radiologists, for the performance of Deep Learning methods. First, we use bone suppression, an algorithm to artificially remove the rib cage. Secondly, we employ an automatic lung field detection to crop the image to the lung area. Furthermore, we consider the combination of both in the context of an ensemble approach. In a five-times re-sampling scheme, we use Receiver Operating Characteristic (ROC) statistics to evaluate the effect of the pre-processing approaches. Using a Convolutional Neural Network (CNN), optimized for X-ray analysis, we achieve a good performance with respect to all pathologies on average. Superior results are obtained for selected pathologies when using pre-processing, i.e. for mass the area under the ROC curve increased by 9.95%. The ensemble with pre-processed trained models yields the best overall results.
Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification
Mar 06, 2018
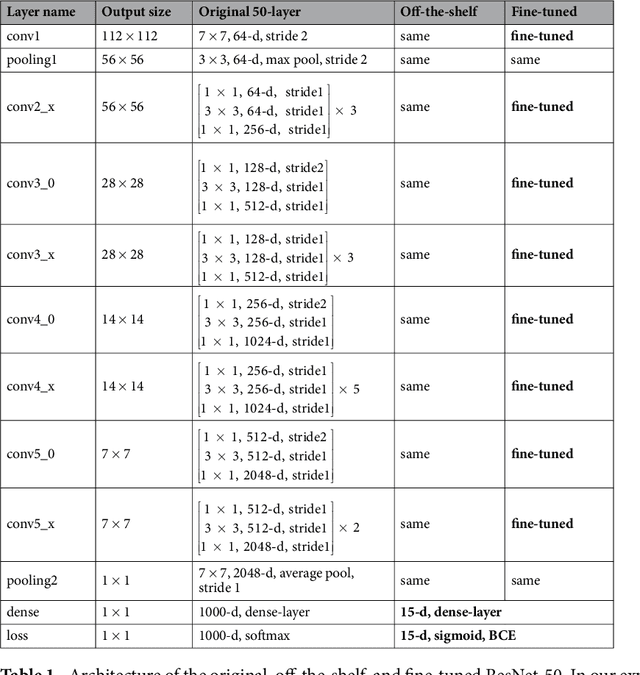
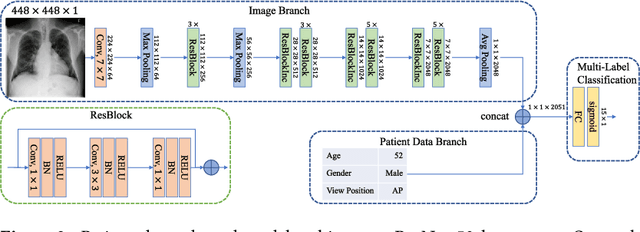
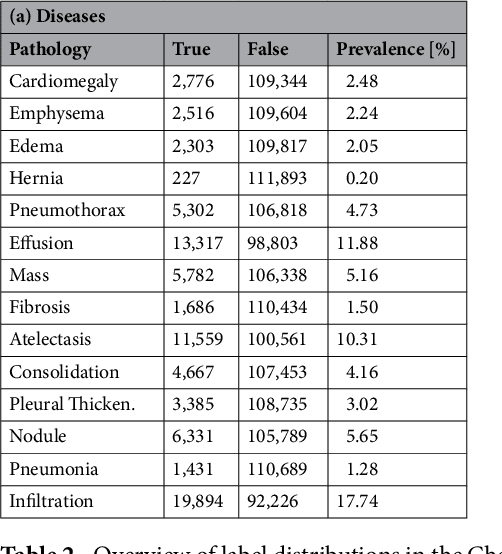
Abstract:The increased availability of X-ray image archives (e.g. the ChestX-ray14 dataset from the NIH Clinical Center) has triggered a growing interest in deep learning techniques. To provide better insight into the different approaches, and their applications to chest X-ray classification, we investigate a powerful network architecture in detail: the ResNet-50. Building on prior work in this domain, we consider transfer learning with and without fine-tuning as well as the training of a dedicated X-ray network from scratch. To leverage the high spatial resolutions of X-ray data, we also include an extended ResNet-50 architecture, and a network integrating non-image data (patient age, gender and acquisition type) in the classification process. In a systematic evaluation, using 5-fold re-sampling and a multi-label loss function, we evaluate the performance of the different approaches for pathology classification by ROC statistics and analyze differences between the classifiers using rank correlation. We observe a considerable spread in the achieved performance and conclude that the X-ray-specific ResNet-50, integrating non-image data yields the best overall results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge