Jens von Berg
When does Bone Suppression and Lung Field Segmentation Improve Chest X-Ray Disease Classification?
Oct 17, 2018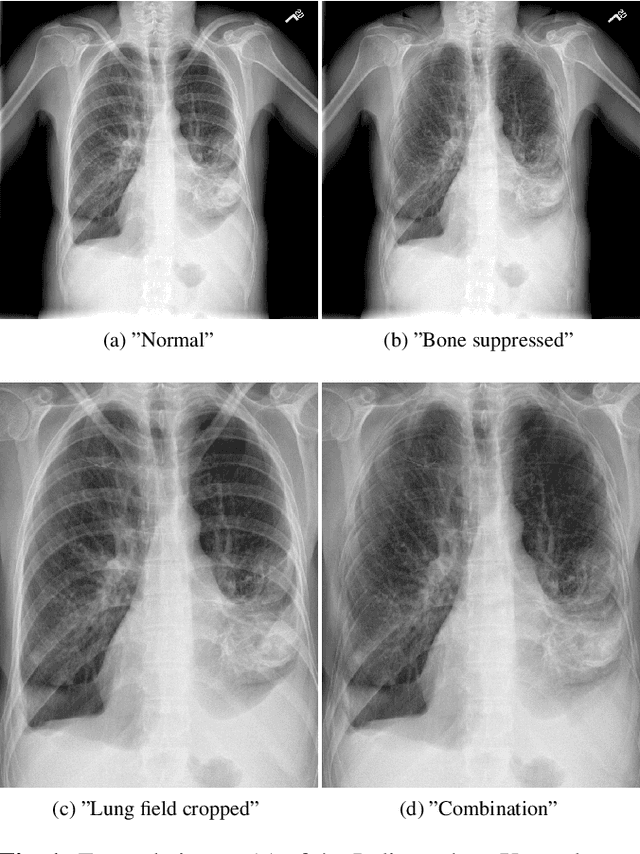
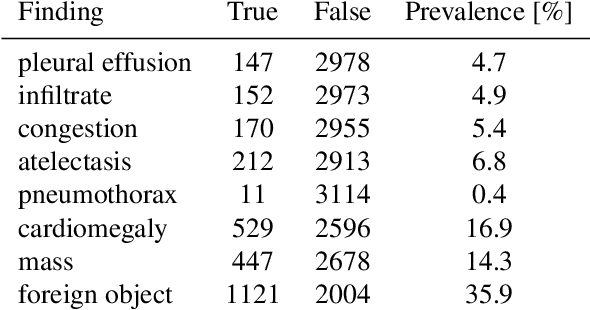
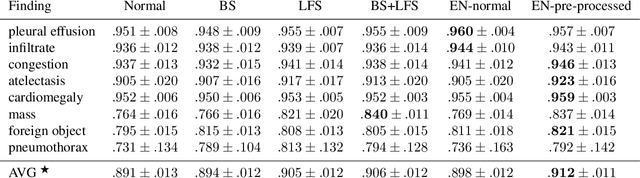
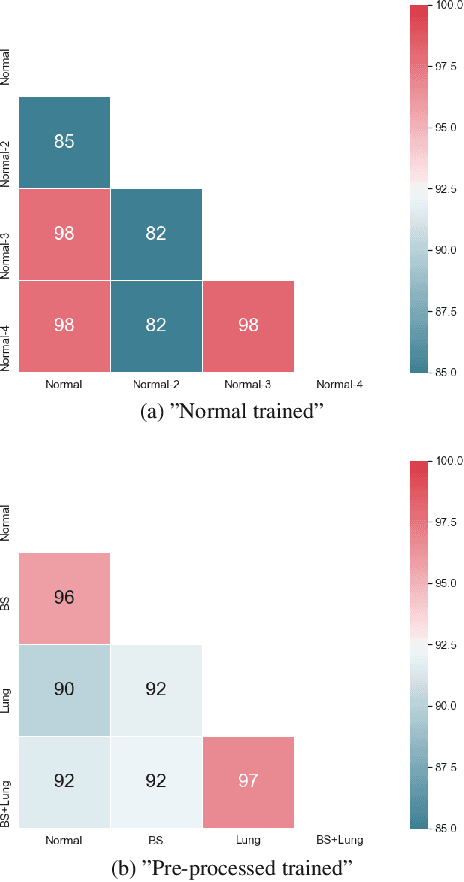
Abstract:Chest radiography is the most common clinical examination type. To improve the quality of patient care and to reduce workload, methods for automatic pathology classification have been developed. In this contribution we investigate the usefulness of two advanced image pre-processing techniques, initially developed for image reading by radiologists, for the performance of Deep Learning methods. First, we use bone suppression, an algorithm to artificially remove the rib cage. Secondly, we employ an automatic lung field detection to crop the image to the lung area. Furthermore, we consider the combination of both in the context of an ensemble approach. In a five-times re-sampling scheme, we use Receiver Operating Characteristic (ROC) statistics to evaluate the effect of the pre-processing approaches. Using a Convolutional Neural Network (CNN), optimized for X-ray analysis, we achieve a good performance with respect to all pathologies on average. Superior results are obtained for selected pathologies when using pre-processing, i.e. for mass the area under the ROC curve increased by 9.95%. The ensemble with pre-processed trained models yields the best overall results.
Deep Learning Based Rib Centerline Extraction and Labeling
Sep 19, 2018



Abstract:Automated extraction and labeling of rib centerlines is a typically needed prerequisite for more advanced assisted reading tools that help the radiologist to efficiently inspect all 24 ribs in a CT volume. In this paper, we combine a deep learning-based rib detection with a dedicated centerline extraction algorithm applied to the detection result for the purpose of fast, robust and accurate rib centerline extraction and labeling from CT volumes. More specifically, we first apply a fully convolutional neural network (FCNN) to generate a probability map for detecting the first rib pair, the twelfth rib pair, and the collection of all intermediate ribs. In a second stage, a newly designed centerline extraction algorithm is applied to this multi-label probability map. Finally, the distinct detection of first and twelfth rib separately, allows to derive individual rib labels by simple sorting and counting the detected centerlines. We applied our method to CT volumes from 116 patients which included a variety of different challenges and achieved a centerline accuracy of 0.787 mm with respect to manual centerline annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge