Fàtima Crispi
Unsupervised Segmentation of Fetal Brain MRI using Deep Learning Cascaded Registration
Jul 07, 2023



Abstract:Accurate segmentation of fetal brain magnetic resonance images is crucial for analyzing fetal brain development and detecting potential neurodevelopmental abnormalities. Traditional deep learning-based automatic segmentation, although effective, requires extensive training data with ground-truth labels, typically produced by clinicians through a time-consuming annotation process. To overcome this challenge, we propose a novel unsupervised segmentation method based on multi-atlas segmentation, that accurately segments multiple tissues without relying on labeled data for training. Our method employs a cascaded deep learning network for 3D image registration, which computes small, incremental deformations to the moving image to align it precisely with the fixed image. This cascaded network can then be used to register multiple annotated images with the image to be segmented, and combine the propagated labels to form a refined segmentation. Our experiments demonstrate that the proposed cascaded architecture outperforms the state-of-the-art registration methods that were tested. Furthermore, the derived segmentation method achieves similar performance and inference time to nnU-Net while only using a small subset of annotated data for the multi-atlas segmentation task and none for training the network. Our pipeline for registration and multi-atlas segmentation is publicly available at https://github.com/ValBcn/CasReg.
Handling confounding variables in statistical shape analysis -- application to cardiac remodelling
Jul 28, 2020
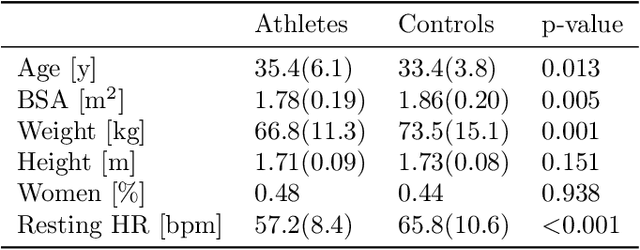
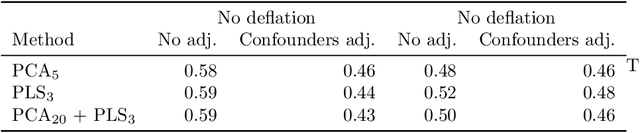
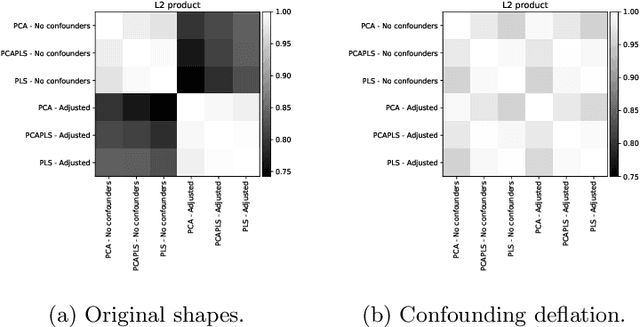
Abstract:Statistical shape analysis is a powerful tool to assess organ morphologies and find shape changes associated to a particular disease. However, imbalance in confounding factors, such as demographics might invalidate the analysis if not taken into consideration. Despite the methodological advances in the field, providing new methods that are able to capture complex and regional shape differences, the relationship between non-imaging information and shape variability has been overlooked. We present a linear statistical shape analysis framework that finds shape differences unassociated to a controlled set of confounding variables. It includes two confounding correction methods: confounding deflation and adjustment. We applied our framework to a cardiac magnetic resonance imaging dataset, consisting of the cardiac ventricles of 89 triathletes and 77 controls, to identify cardiac remodelling due to the practice of endurance exercise. To test robustness to confounders, subsets of this dataset were generated by randomly removing controls with low body mass index, thus introducing imbalance. The analysis of the whole dataset indicates an increase of ventricular volumes and myocardial mass in athletes, which is consistent with the clinical literature. However, when confounders are not taken into consideration no increase of myocardial mass is found. Using the downsampled datasets, we find that confounder adjustment methods are needed to find the real remodelling patterns in imbalanced datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge