Miguel Ángel González Ballester
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025



Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
AIROGS: Artificial Intelligence for RObust Glaucoma Screening Challenge
Feb 10, 2023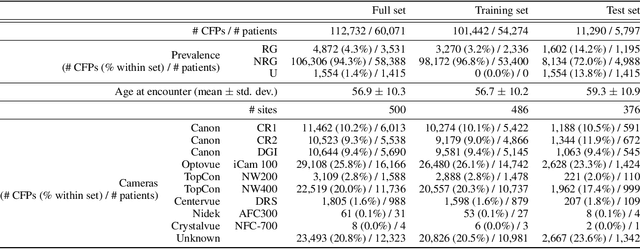
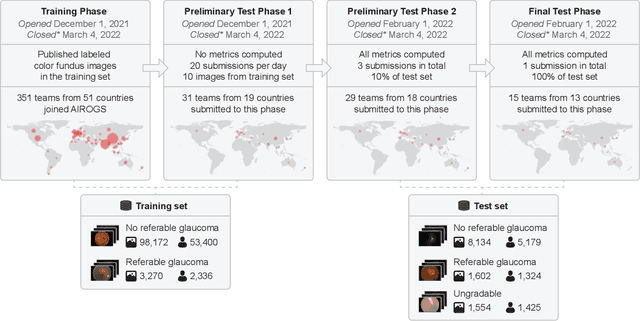
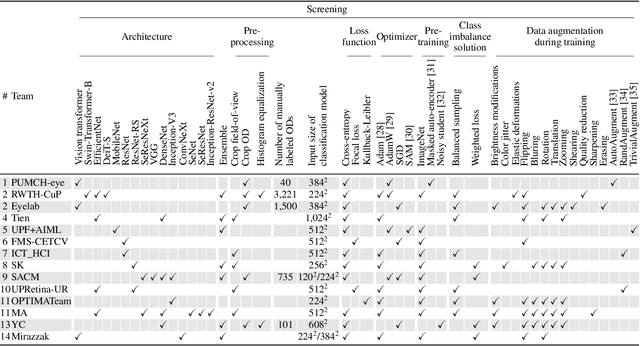
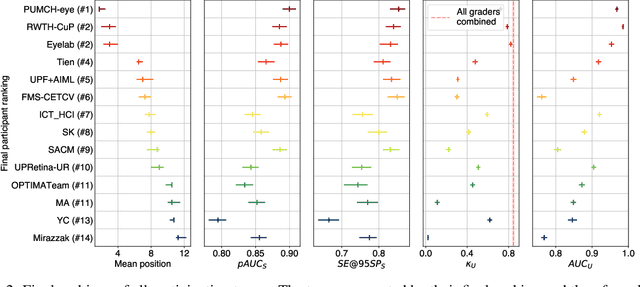
Abstract:The early detection of glaucoma is essential in preventing visual impairment. Artificial intelligence (AI) can be used to analyze color fundus photographs (CFPs) in a cost-effective manner, making glaucoma screening more accessible. While AI models for glaucoma screening from CFPs have shown promising results in laboratory settings, their performance decreases significantly in real-world scenarios due to the presence of out-of-distribution and low-quality images. To address this issue, we propose the Artificial Intelligence for Robust Glaucoma Screening (AIROGS) challenge. This challenge includes a large dataset of around 113,000 images from about 60,000 patients and 500 different screening centers, and encourages the development of algorithms that are robust to ungradable and unexpected input data. We evaluated solutions from 14 teams in this paper, and found that the best teams performed similarly to a set of 20 expert ophthalmologists and optometrists. The highest-scoring team achieved an area under the receiver operating characteristic curve of 0.99 (95% CI: 0.98-0.99) for detecting ungradable images on-the-fly. Additionally, many of the algorithms showed robust performance when tested on three other publicly available datasets. These results demonstrate the feasibility of robust AI-enabled glaucoma screening.
On the Optimal Combination of Cross-Entropy and Soft Dice Losses for Lesion Segmentation with Out-of-Distribution Robustness
Sep 14, 2022


Abstract:We study the impact of different loss functions on lesion segmentation from medical images. Although the Cross-Entropy (CE) loss is the most popular option when dealing with natural images, for biomedical image segmentation the soft Dice loss is often preferred due to its ability to handle imbalanced scenarios. On the other hand, the combination of both functions has also been successfully applied in this kind of tasks. A much less studied problem is the generalization ability of all these losses in the presence of Out-of-Distribution (OoD) data. This refers to samples appearing in test time that are drawn from a different distribution than training images. In our case, we train our models on images that always contain lesions, but in test time we also have lesion-free samples. We analyze the impact of the minimization of different loss functions on in-distribution performance, but also its ability to generalize to OoD data, via comprehensive experiments on polyp segmentation from endoscopic images and ulcer segmentation from diabetic feet images. Our findings are surprising: CE-Dice loss combinations that excel in segmenting in-distribution images have a poor performance when dealing with OoD data, which leads us to recommend the adoption of the CE loss for this kind of problems, due to its robustness and ability to generalize to OoD samples. Code associated to our experiments can be found at https://github.com/agaldran/lesion_losses_ood .
Handling confounding variables in statistical shape analysis -- application to cardiac remodelling
Jul 28, 2020
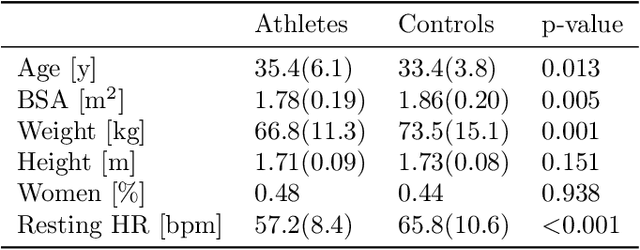
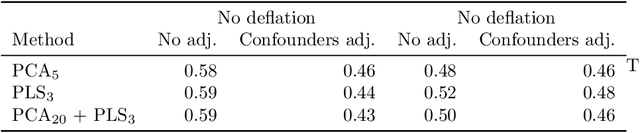
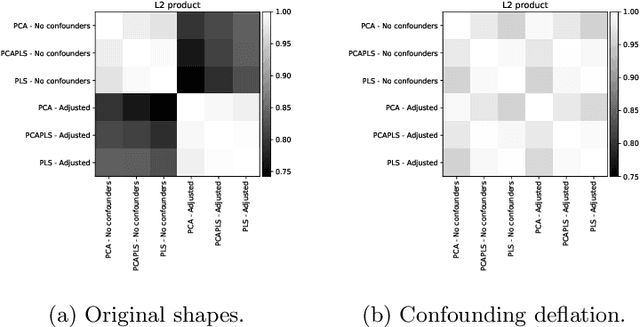
Abstract:Statistical shape analysis is a powerful tool to assess organ morphologies and find shape changes associated to a particular disease. However, imbalance in confounding factors, such as demographics might invalidate the analysis if not taken into consideration. Despite the methodological advances in the field, providing new methods that are able to capture complex and regional shape differences, the relationship between non-imaging information and shape variability has been overlooked. We present a linear statistical shape analysis framework that finds shape differences unassociated to a controlled set of confounding variables. It includes two confounding correction methods: confounding deflation and adjustment. We applied our framework to a cardiac magnetic resonance imaging dataset, consisting of the cardiac ventricles of 89 triathletes and 77 controls, to identify cardiac remodelling due to the practice of endurance exercise. To test robustness to confounders, subsets of this dataset were generated by randomly removing controls with low body mass index, thus introducing imbalance. The analysis of the whole dataset indicates an increase of ventricular volumes and myocardial mass in athletes, which is consistent with the clinical literature. However, when confounders are not taken into consideration no increase of myocardial mass is found. Using the downsampled datasets, we find that confounder adjustment methods are needed to find the real remodelling patterns in imbalanced datasets.
Re-Identification and Growth Detection of Pulmonary Nodules without Image Registration Using 3D Siamese Neural Networks
Dec 22, 2019
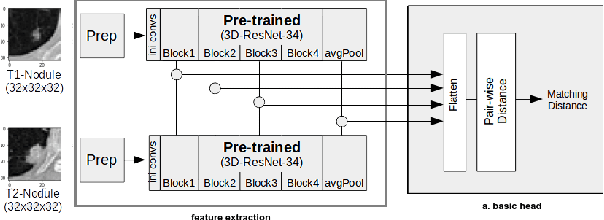
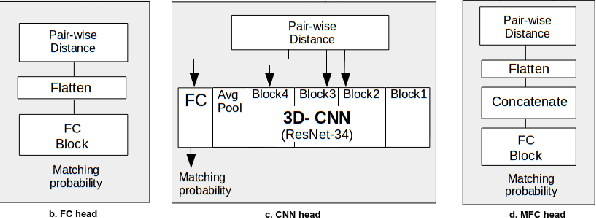
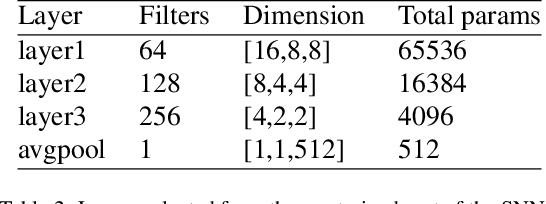
Abstract:Lung cancer follow-up is a complex, error prone, and time consuming task for clinical radiologists. Several lung CT scan images taken at different time points of a given patient need to be individually inspected, looking for possible cancerogenous nodules. Radiologists mainly focus their attention in nodule size, density, and growth to assess the existence of malignancy. In this study, we present a novel method based on a 3D siamese neural network, for the re-identification of nodules in a pair of CT scans of the same patient without the need for image registration. The network was integrated into a two-stage automatic pipeline to detect, match, and predict nodule growth given pairs of CT scans. Results on an independent test set reported a nodule detection sensitivity of 94.7%, an accuracy for temporal nodule matching of 88.8%, and a sensitivity of 92.0% with a precision of 88.4% for nodule growth detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge