Gemma Piella
BCN MedTech, Dept. of Information and Communication Technologies, Universitat Pompeu Fabra, Barcelona, Spain
BabyFlow: 3D modeling of realistic and expressive infant faces
Dec 22, 2025Abstract:Early detection of developmental disorders can be aided by analyzing infant craniofacial morphology, but modeling infant faces is challenging due to limited data and frequent spontaneous expressions. We introduce BabyFlow, a generative AI model that disentangles facial identity and expression, enabling independent control over both. Using normalizing flows, BabyFlow learns flexible, probabilistic representations that capture the complex, non-linear variability of expressive infant faces without restrictive linear assumptions. To address scarce and uncontrolled expressive data, we perform cross-age expression transfer, adapting expressions from adult 3D scans to enrich infant datasets with realistic and systematic expressive variants. As a result, BabyFlow improves 3D reconstruction accuracy, particularly in highly expressive regions such as the mouth, eyes, and nose, and supports synthesis and modification of infant expressions while preserving identity. Additionally, by integrating with diffusion models, BabyFlow generates high-fidelity 2D infant images with consistent 3D geometry, providing powerful tools for data augmentation and early facial analysis.
Fetpype: An Open-Source Pipeline for Reproducible Fetal Brain MRI Analysis
Dec 19, 2025Abstract:Fetal brain Magnetic Resonance Imaging (MRI) is crucial for assessing neurodevelopment in utero. However, analyzing this data presents significant challenges due to fetal motion, low signal-to-noise ratio, and the need for complex multi-step processing, including motion correction, super-resolution reconstruction, segmentation, and surface extraction. While various specialized tools exist for individual steps, integrating them into robust, reproducible, and user-friendly workflows that go from raw images to processed volumes is not straightforward. This lack of standardization hinders reproducibility across studies and limits the adoption of advanced analysis techniques for researchers and clinicians. To address these challenges, we introduce Fetpype, an open-source Python library designed to streamline and standardize the preprocessing and analysis of T2-weighted fetal brain MRI data. Fetpype is publicly available on GitHub at https://github.com/fetpype/fetpype.
Automatic facial axes standardization of 3D fetal ultrasound images
Sep 04, 2024Abstract:Craniofacial anomalies indicate early developmental disturbances and are usually linked to many genetic syndromes. Early diagnosis is critical, yet ultrasound (US) examinations often fail to identify these features. This study presents an AI-driven tool to assist clinicians in standardizing fetal facial axes/planes in 3D US, reducing sonographer workload and facilitating the facial evaluation. Our network, structured into three blocks-feature extractor, rotation and translation regression, and spatial transformer-processes three orthogonal 2D slices to estimate the necessary transformations for standardizing the facial planes in the 3D US. These transformations are applied to the original 3D US using a differentiable module (the spatial transformer block), yielding a standardized 3D US and the corresponding 2D facial standard planes. The dataset used consists of 1180 fetal facial 3D US images acquired between weeks 20 and 35 of gestation. Results show that our network considerably reduces inter-observer rotation variability in the test set, with a mean geodesic angle difference of 14.12$^{\circ}$ $\pm$ 18.27$^{\circ}$ and an Euclidean angle error of 7.45$^{\circ}$ $\pm$ 14.88$^{\circ}$. These findings demonstrate the network's ability to effectively standardize facial axes, crucial for consistent fetal facial assessments. In conclusion, the proposed network demonstrates potential for improving the consistency and accuracy of fetal facial assessments in clinical settings, facilitating early evaluation of craniofacial anomalies.
Unsupervised Segmentation of Fetal Brain MRI using Deep Learning Cascaded Registration
Jul 07, 2023



Abstract:Accurate segmentation of fetal brain magnetic resonance images is crucial for analyzing fetal brain development and detecting potential neurodevelopmental abnormalities. Traditional deep learning-based automatic segmentation, although effective, requires extensive training data with ground-truth labels, typically produced by clinicians through a time-consuming annotation process. To overcome this challenge, we propose a novel unsupervised segmentation method based on multi-atlas segmentation, that accurately segments multiple tissues without relying on labeled data for training. Our method employs a cascaded deep learning network for 3D image registration, which computes small, incremental deformations to the moving image to align it precisely with the fixed image. This cascaded network can then be used to register multiple annotated images with the image to be segmented, and combine the propagated labels to form a refined segmentation. Our experiments demonstrate that the proposed cascaded architecture outperforms the state-of-the-art registration methods that were tested. Furthermore, the derived segmentation method achieves similar performance and inference time to nnU-Net while only using a small subset of annotated data for the multi-atlas segmentation task and none for training the network. Our pipeline for registration and multi-atlas segmentation is publicly available at https://github.com/ValBcn/CasReg.
BabyNet: Reconstructing 3D faces of babies from uncalibrated photographs
Mar 11, 2022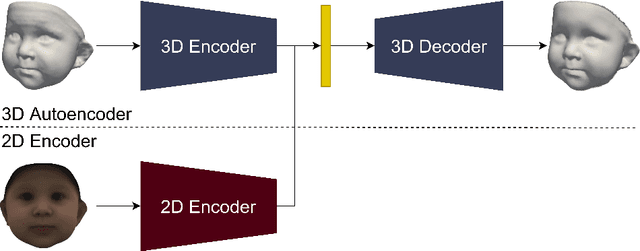
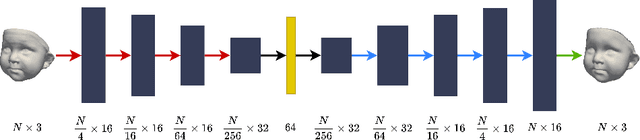
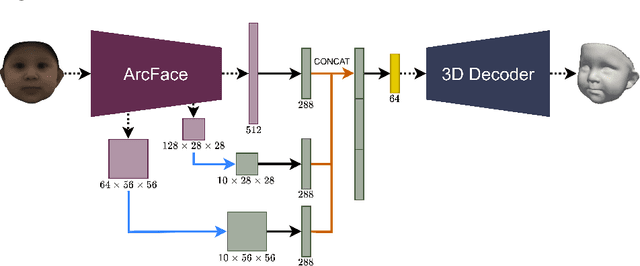
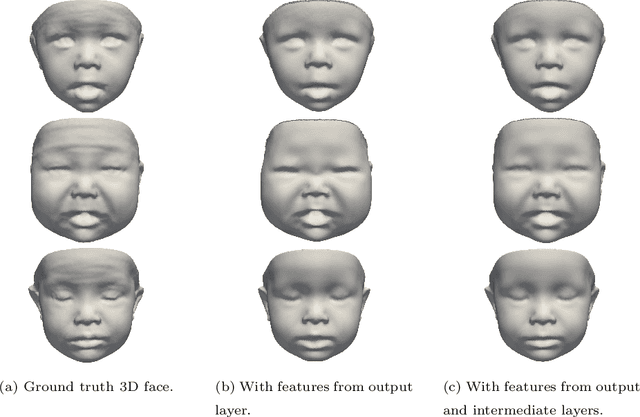
Abstract:We present a 3D face reconstruction system that aims at recovering the 3D facial geometry of babies from uncalibrated photographs, BabyNet. Since the 3D facial geometry of babies differs substantially from that of adults, baby-specific facial reconstruction systems are needed. BabyNet consists of two stages: 1) a 3D graph convolutional autoencoder learns a latent space of the baby 3D facial shape; and 2) a 2D encoder that maps photographs to the 3D latent space based on representative features extracted using transfer learning. In this way, using the pre-trained 3D decoder, we can recover a 3D face from 2D images. We evaluate BabyNet and show that 1) methods based on adult datasets cannot model the 3D facial geometry of babies, which proves the need for a baby-specific method, and 2) BabyNet outperforms classical model-fitting methods even when a baby-specific 3D morphable model, such as BabyFM, is used.
Memory-aware curriculum federated learning for breast cancer classification
Jul 06, 2021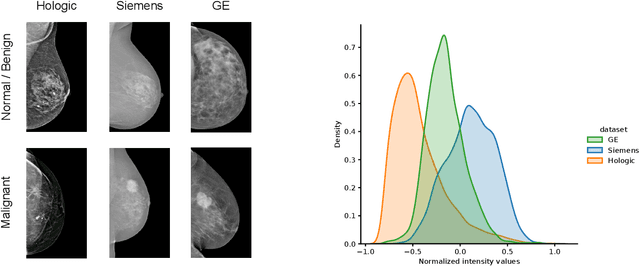
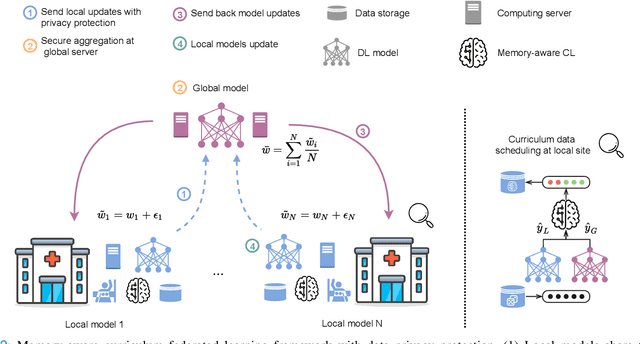
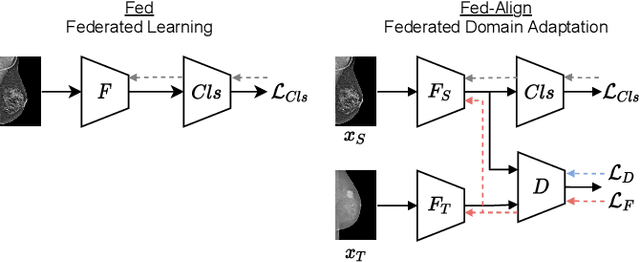
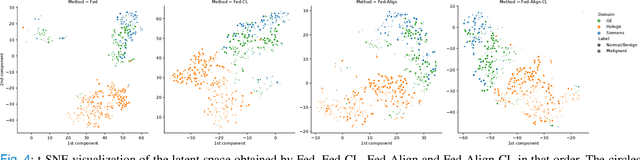
Abstract:For early breast cancer detection, regular screening with mammography imaging is recommended. Routinary examinations result in datasets with a predominant amount of negative samples. A potential solution to such class-imbalance is joining forces across multiple institutions. Developing a collaborative computer-aided diagnosis system is challenging in different ways. Patient privacy and regulations need to be carefully respected. Data across institutions may be acquired from different devices or imaging protocols, leading to heterogeneous non-IID data. Also, for learning-based methods, new optimization strategies working on distributed data are required. Recently, federated learning has emerged as an effective tool for collaborative learning. In this setting, local models perform computation on their private data to update the global model. The order and the frequency of local updates influence the final global model. Hence, the order in which samples are locally presented to the optimizers plays an important role. In this work, we define a memory-aware curriculum learning method for the federated setting. Our curriculum controls the order of the training samples paying special attention to those that are forgotten after the deployment of the global model. Our approach is combined with unsupervised domain adaptation to deal with domain shift while preserving data privacy. We evaluate our method with three clinical datasets from different vendors. Our results verify the effectiveness of federated adversarial learning for the multi-site breast cancer classification. Moreover, we show that our proposed memory-aware curriculum method is beneficial to further improve classification performance. Our code is publicly available at: https://github.com/ameliajimenez/curriculum-federated-learning.
An Uncertainty-aware Hierarchical Probabilistic Network for Early Prediction, Quantification and Segmentation of Pulmonary Tumour Growth
Apr 18, 2021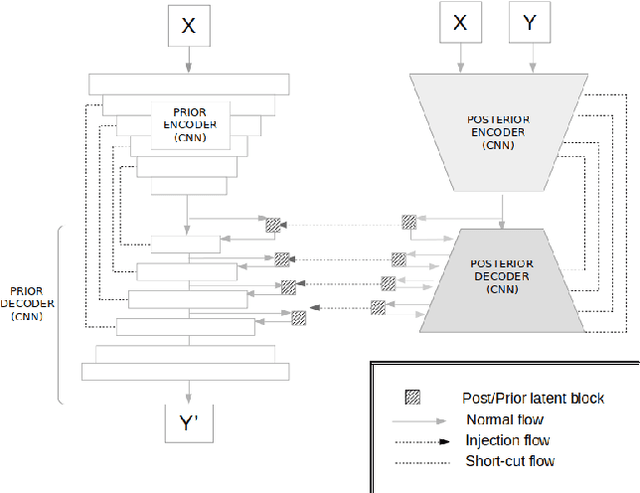
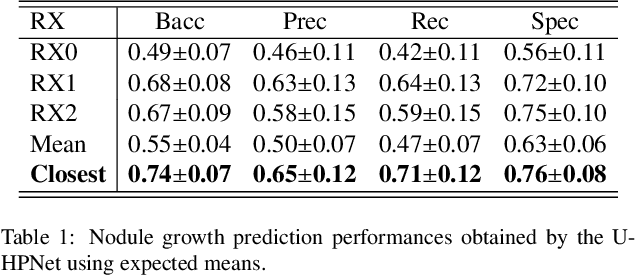
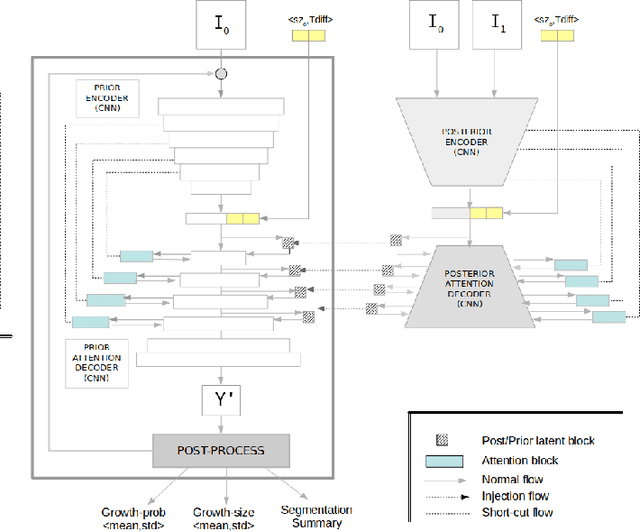
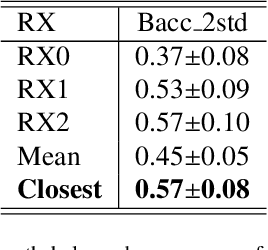
Abstract:Early detection and quantification of tumour growth would help clinicians to prescribe more accurate treatments and provide better surgical planning. However, the multifactorial and heterogeneous nature of lung tumour progression hampers identification of growth patterns. In this study, we present a novel method based on a deep hierarchical generative and probabilistic framework that, according to radiological guidelines, predicts tumour growth, quantifies its size and provides a semantic appearance of the future nodule. Unlike previous deterministic solutions, the generative characteristic of our approach also allows us to estimate the uncertainty in the predictions, especially important for complex and doubtful cases. Results of evaluating this method on an independent test set reported a tumour growth balanced accuracy of 74%, a tumour growth size MAE of 1.77 mm and a tumour segmentation Dice score of 78%. These surpassed the performances of equivalent deterministic and alternative generative solutions (i.e. probabilistic U-Net, Bayesian test dropout and Pix2Pix GAN) confirming the suitability of our approach.
Detection, growth quantification and malignancy prediction of pulmonary nodules using deep convolutional networks in follow-up CT scans
Mar 26, 2021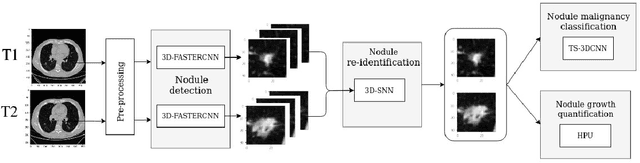
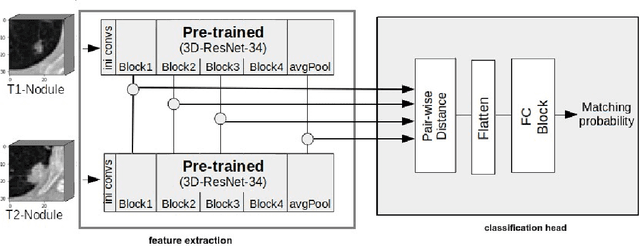
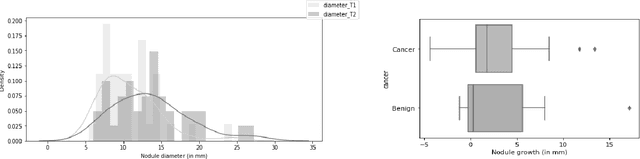
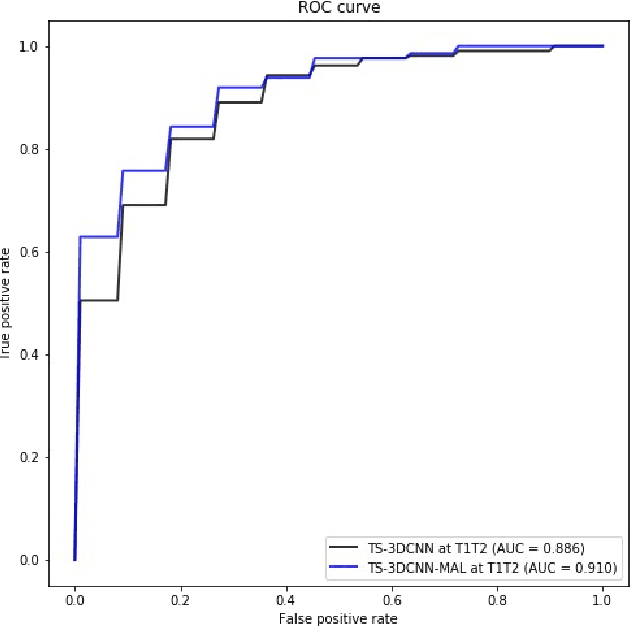
Abstract:We address the problem of supporting radiologists in the longitudinal management of lung cancer. Therefore, we proposed a deep learning pipeline, composed of four stages that completely automatized from the detection of nodules to the classification of cancer, through the detection of growth in the nodules. In addition, the pipeline integrated a novel approach for nodule growth detection, which relied on a recent hierarchical probabilistic U-Net adapted to report uncertainty estimates. Also, a second novel method was introduced for lung cancer nodule classification, integrating into a two stream 3D-CNN network the estimated nodule malignancy probabilities derived from a pretrained nodule malignancy network. The pipeline was evaluated in a longitudinal cohort and reported comparable performances to the state of art.
Survey on 3D face reconstruction from uncalibrated images
Nov 11, 2020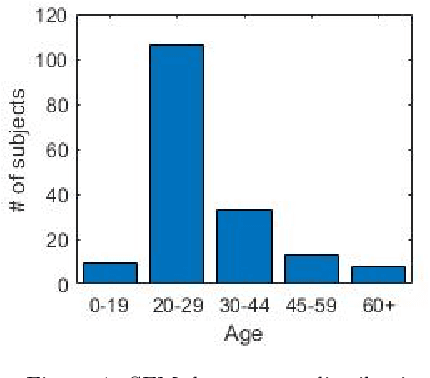
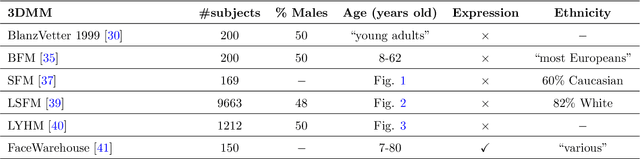

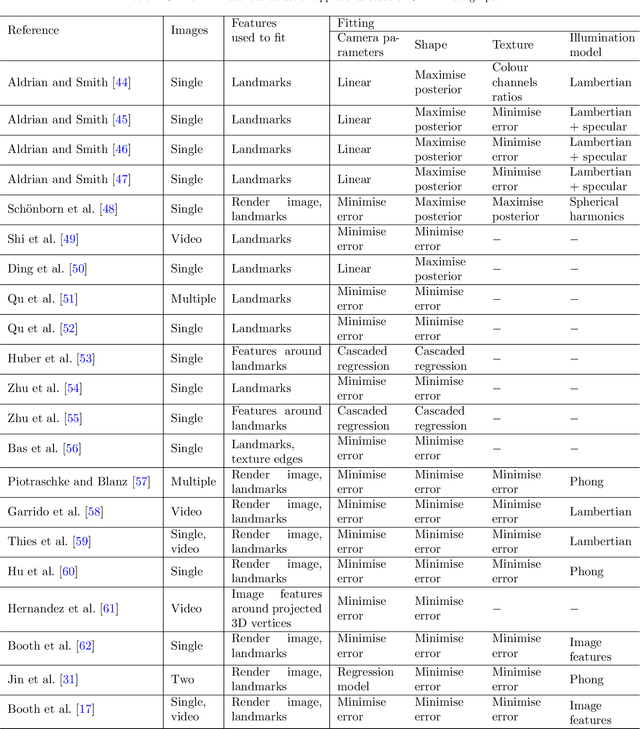
Abstract:Recently, a lot of attention has been focused on the incorporation of 3D data into face analysis and its applications. Despite providing a more accurate representation of the face, 3D face images are more complex to acquire than 2D pictures. As a consequence, great effort has been invested in developing systems that reconstruct 3D faces from an uncalibrated 2D image. However, the 3D-from-2D face reconstruction problem is ill-posed, thus prior knowledge is needed to restrict the solutions space. In this work, we review 3D face reconstruction methods in the last decade, focusing on those that only use 2D pictures captured under uncontrolled conditions. We present a classification of the proposed methods based on the technique used to add prior knowledge, considering three main strategies, namely, statistical model fitting, photometry, and deep learning, and reviewing each of them separately. In addition, given the relevance of statistical 3D facial models as prior knowledge, we explain the construction procedure and provide a comprehensive list of the publicly available 3D facial models. After the exhaustive study of 3D-from-2D face reconstruction approaches, we observe that the deep learning strategy is rapidly growing since the last few years, matching its extension to that of the widespread statistical model fitting. Unlike the other two strategies, photometry-based methods have decreased in number since the required strong assumptions cause the reconstructions to be of more limited quality than those resulting from model fitting and deep learning methods. The review also identifies current gaps and suggests avenues for future research.
Curriculum learning for annotation-efficient medical image analysis: scheduling data with prior knowledge and uncertainty
Jul 31, 2020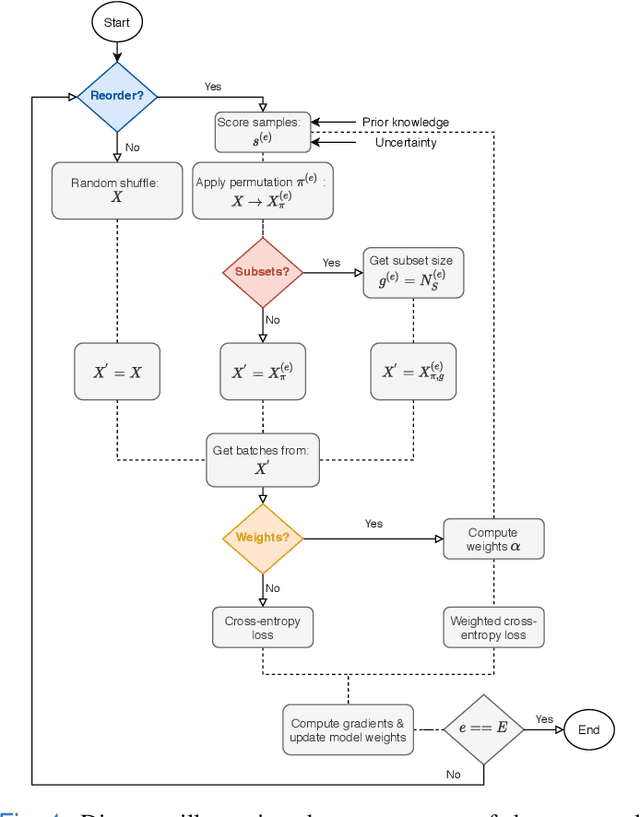
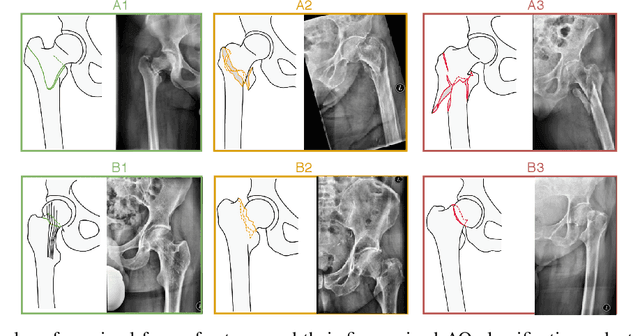
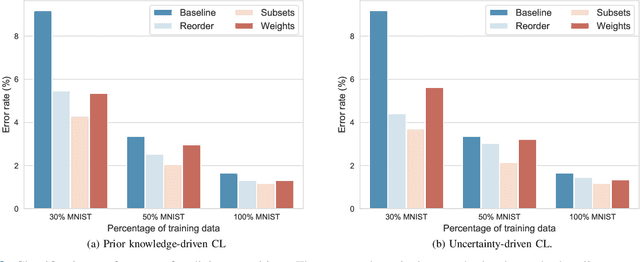
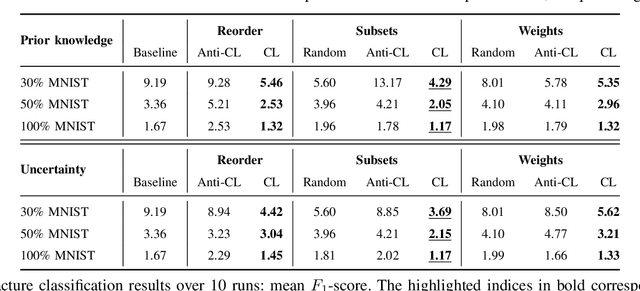
Abstract:Convolutional neural networks (CNNs) for multi-class classification require training on large, representative, and high quality annotated datasets. However, in the field of medical imaging, data and annotations are both difficult and expensive to acquire. Moreover, they frequently suffer from highly imbalanced distributions, and potentially noisy labels due to intra- or inter-expert disagreement. To deal with such challenges, we propose a unified curriculum learning framework to schedule the order and pace of the training samples presented to the optimizer. Our novel framework reunites three strategies consisting of individually weighting training samples, reordering the training set, or sampling subsets of data. The core of these strategies is a scoring function ranking the training samples according to either difficulty or uncertainty. We define the scoring function from domain-specific prior knowledge or by directly measuring the uncertainty in the predictions. We perform a variety of experiments with a clinical dataset for the multi-class classification of proximal femur fractures and the publicly available MNIST dataset. Our results show that the sequence and weight of the training samples play an important role in the optimization process of CNNs. Proximal femur fracture classification is improved up to the performance of experienced trauma surgeons. We further demonstrate the benefits of our unified curriculum learning method for three controlled and challenging digit recognition scenarios: with limited amounts of data, under class-imbalance, and in the presence of label noise.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge