Antonia Alomar
FeTTL: Federated Template and Task Learning for Multi-Institutional Medical Imaging
Jan 22, 2026Abstract:Federated learning enables collaborative model training across geographically distributed medical centers while preserving data privacy. However, domain shifts and heterogeneity in data often lead to a degradation in model performance. Medical imaging applications are particularly affected by variations in acquisition protocols, scanner types, and patient populations. To address these issues, we introduce Federated Template and Task Learning (FeTTL), a novel framework designed to harmonize multi-institutional medical imaging data in federated environments. FeTTL learns a global template together with a task model to align data distributions among clients. We evaluated FeTTL on two challenging and diverse multi-institutional medical imaging tasks: retinal fundus optical disc segmentation and histopathological metastasis classification. Experimental results show that FeTTL significantly outperforms the state-of-the-art federated learning baselines (p-values <0.002) for optical disc segmentation and classification of metastases from multi-institutional data. Our experiments further highlight the importance of jointly learning the template and the task. These findings suggest that FeTTL offers a principled and extensible solution for mitigating distribution shifts in federated learning, supporting robust model deployment in real-world, multi-institutional environments.
BabyFlow: 3D modeling of realistic and expressive infant faces
Dec 22, 2025Abstract:Early detection of developmental disorders can be aided by analyzing infant craniofacial morphology, but modeling infant faces is challenging due to limited data and frequent spontaneous expressions. We introduce BabyFlow, a generative AI model that disentangles facial identity and expression, enabling independent control over both. Using normalizing flows, BabyFlow learns flexible, probabilistic representations that capture the complex, non-linear variability of expressive infant faces without restrictive linear assumptions. To address scarce and uncontrolled expressive data, we perform cross-age expression transfer, adapting expressions from adult 3D scans to enrich infant datasets with realistic and systematic expressive variants. As a result, BabyFlow improves 3D reconstruction accuracy, particularly in highly expressive regions such as the mouth, eyes, and nose, and supports synthesis and modification of infant expressions while preserving identity. Additionally, by integrating with diffusion models, BabyFlow generates high-fidelity 2D infant images with consistent 3D geometry, providing powerful tools for data augmentation and early facial analysis.
Automatic facial axes standardization of 3D fetal ultrasound images
Sep 04, 2024Abstract:Craniofacial anomalies indicate early developmental disturbances and are usually linked to many genetic syndromes. Early diagnosis is critical, yet ultrasound (US) examinations often fail to identify these features. This study presents an AI-driven tool to assist clinicians in standardizing fetal facial axes/planes in 3D US, reducing sonographer workload and facilitating the facial evaluation. Our network, structured into three blocks-feature extractor, rotation and translation regression, and spatial transformer-processes three orthogonal 2D slices to estimate the necessary transformations for standardizing the facial planes in the 3D US. These transformations are applied to the original 3D US using a differentiable module (the spatial transformer block), yielding a standardized 3D US and the corresponding 2D facial standard planes. The dataset used consists of 1180 fetal facial 3D US images acquired between weeks 20 and 35 of gestation. Results show that our network considerably reduces inter-observer rotation variability in the test set, with a mean geodesic angle difference of 14.12$^{\circ}$ $\pm$ 18.27$^{\circ}$ and an Euclidean angle error of 7.45$^{\circ}$ $\pm$ 14.88$^{\circ}$. These findings demonstrate the network's ability to effectively standardize facial axes, crucial for consistent fetal facial assessments. In conclusion, the proposed network demonstrates potential for improving the consistency and accuracy of fetal facial assessments in clinical settings, facilitating early evaluation of craniofacial anomalies.
PhysFlow: Skin tone transfer for remote heart rate estimation through conditional normalizing flows
Jul 31, 2024Abstract:In recent years, deep learning methods have shown impressive results for camera-based remote physiological signal estimation, clearly surpassing traditional methods. However, the performance and generalization ability of Deep Neural Networks heavily depends on rich training data truly representing different factors of variation encountered in real applications. Unfortunately, many current remote photoplethysmography (rPPG) datasets lack diversity, particularly in darker skin tones, leading to biased performance of existing rPPG approaches. To mitigate this bias, we introduce PhysFlow, a novel method for augmenting skin diversity in remote heart rate estimation using conditional normalizing flows. PhysFlow adopts end-to-end training optimization, enabling simultaneous training of supervised rPPG approaches on both original and generated data. Additionally, we condition our model using CIELAB color space skin features directly extracted from the facial videos without the need for skin-tone labels. We validate PhysFlow on publicly available datasets, UCLA-rPPG and MMPD, demonstrating reduced heart rate error, particularly in dark skin tones. Furthermore, we demonstrate its versatility and adaptability across different data-driven rPPG methods.
Data Alchemy: Mitigating Cross-Site Model Variability Through Test Time Data Calibration
Jul 18, 2024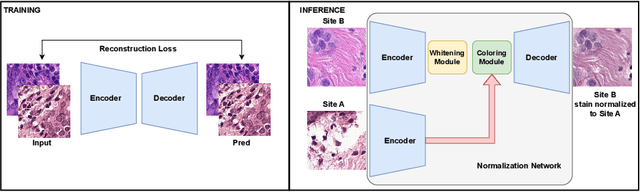
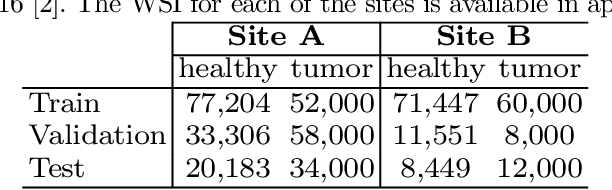
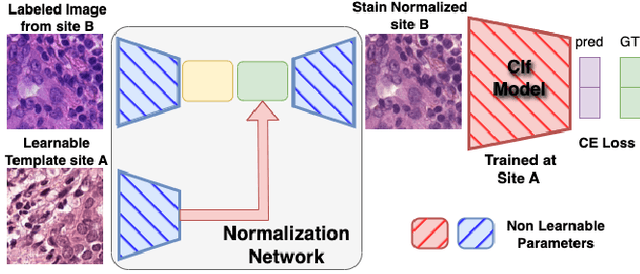
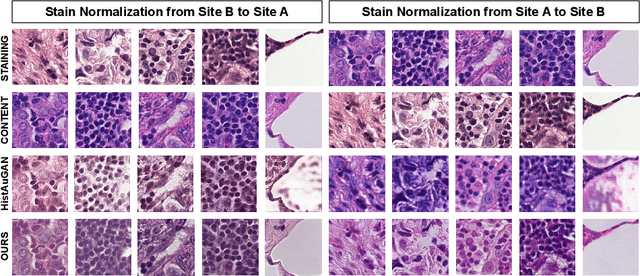
Abstract:Deploying deep learning-based imaging tools across various clinical sites poses significant challenges due to inherent domain shifts and regulatory hurdles associated with site-specific fine-tuning. For histopathology, stain normalization techniques can mitigate discrepancies, but they often fall short of eliminating inter-site variations. Therefore, we present Data Alchemy, an explainable stain normalization method combined with test time data calibration via a template learning framework to overcome barriers in cross-site analysis. Data Alchemy handles shifts inherent to multi-site data and minimizes them without needing to change the weights of the normalization or classifier networks. Our approach extends to unseen sites in various clinical settings where data domain discrepancies are unknown. Extensive experiments highlight the efficacy of our framework in tumor classification in hematoxylin and eosin-stained patches. Our explainable normalization method boosts classification tasks' area under the precision-recall curve(AUPR) by 0.165, 0.545 to 0.710. Additionally, Data Alchemy further reduces the multisite classification domain gap, by improving the 0.710 AUPR an additional 0.142, elevating classification performance further to 0.852, from 0.545. Our Data Alchemy framework can popularize precision medicine with minimal operational overhead by allowing for the seamless integration of pre-trained deep learning-based clinical tools across multiple sites.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge