Abhijeet Parida
Standardized Methods and Recommendations for Green Federated Learning
Jan 30, 2026Abstract:Federated learning (FL) enables collaborative model training over privacy-sensitive, distributed data, but its environmental impact is difficult to compare across studies due to inconsistent measurement boundaries and heterogeneous reporting. We present a practical carbon-accounting methodology for FL CO2e tracking using NVIDIA NVFlare and CodeCarbon for explicit, phase-aware tasks (initialization, per-round training, evaluation, and idle/coordination). To capture non-compute effects, we additionally estimate communication emissions from transmitted model-update sizes under a network-configurable energy model. We validate the proposed approach on two representative workloads: CIFAR-10 image classification and retinal optic disk segmentation. In CIFAR-10, controlled client-efficiency scenarios show that system-level slowdowns and coordination effects can contribute meaningfully to carbon footprint under an otherwise fixed FL protocol, increasing total CO2e by 8.34x (medium) and 21.73x (low) relative to the high-efficiency baseline. In retinal segmentation, swapping GPU tiers (H100 vs.\ V100) yields a consistent 1.7x runtime gap (290 vs. 503 minutes) while producing non-uniform changes in total energy and CO2e across sites, underscoring the need for per-site and per-round reporting. Overall, our results support a standardized carbon accounting method that acts as a prerequisite for reproducible 'green' FL evaluation. Our code is available at https://github.com/Pediatric-Accelerated-Intelligence-Lab/carbon_footprint.
FeTTL: Federated Template and Task Learning for Multi-Institutional Medical Imaging
Jan 22, 2026Abstract:Federated learning enables collaborative model training across geographically distributed medical centers while preserving data privacy. However, domain shifts and heterogeneity in data often lead to a degradation in model performance. Medical imaging applications are particularly affected by variations in acquisition protocols, scanner types, and patient populations. To address these issues, we introduce Federated Template and Task Learning (FeTTL), a novel framework designed to harmonize multi-institutional medical imaging data in federated environments. FeTTL learns a global template together with a task model to align data distributions among clients. We evaluated FeTTL on two challenging and diverse multi-institutional medical imaging tasks: retinal fundus optical disc segmentation and histopathological metastasis classification. Experimental results show that FeTTL significantly outperforms the state-of-the-art federated learning baselines (p-values <0.002) for optical disc segmentation and classification of metastases from multi-institutional data. Our experiments further highlight the importance of jointly learning the template and the task. These findings suggest that FeTTL offers a principled and extensible solution for mitigating distribution shifts in federated learning, supporting robust model deployment in real-world, multi-institutional environments.
Adaptable Segmentation Pipeline for Diverse Brain Tumors with Radiomic-guided Subtyping and Lesion-Wise Model Ensemble
Dec 16, 2025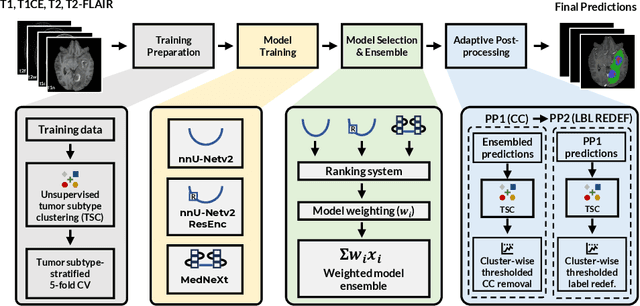
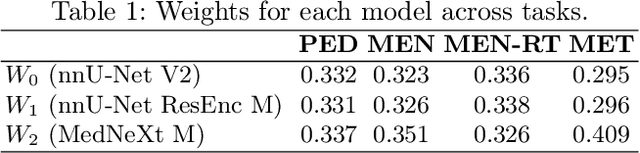
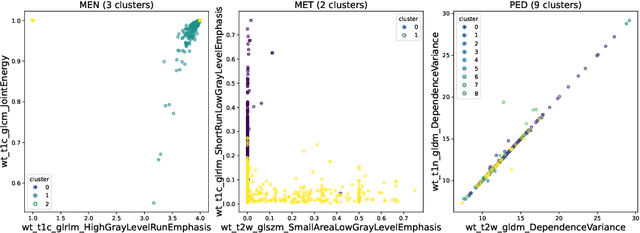
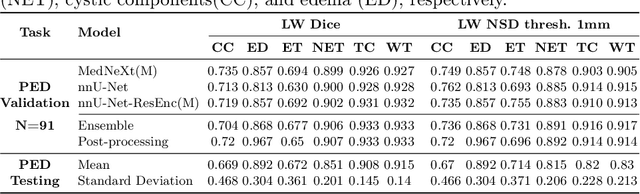
Abstract:Robust and generalizable segmentation of brain tumors on multi-parametric magnetic resonance imaging (MRI) remains difficult because tumor types differ widely. The BraTS 2025 Lighthouse Challenge benchmarks segmentation methods on diverse high-quality datasets of adult and pediatric tumors: multi-consortium international pediatric brain tumor segmentation (PED), preoperative meningioma tumor segmentation (MEN), meningioma radiotherapy segmentation (MEN-RT), and segmentation of pre- and post-treatment brain metastases (MET). We present a flexible, modular, and adaptable pipeline that improves segmentation performance by selecting and combining state-of-the-art models and applying tumor- and lesion-specific processing before and after training. Radiomic features extracted from MRI help detect tumor subtype, ensuring a more balanced training. Custom lesion-level performance metrics determine the influence of each model in the ensemble and optimize post-processing that further refines the predictions, enabling the workflow to tailor every step to each case. On the BraTS testing sets, our pipeline achieved performance comparable to top-ranked algorithms across multiple challenges. These findings confirm that custom lesion-aware processing and model selection yield robust segmentations yet without locking the method to a specific network architecture. Our method has the potential for quantitative tumor measurement in clinical practice, supporting diagnosis and prognosis.
Improving Pre-trained Segmentation Models using Post-Processing
Dec 16, 2025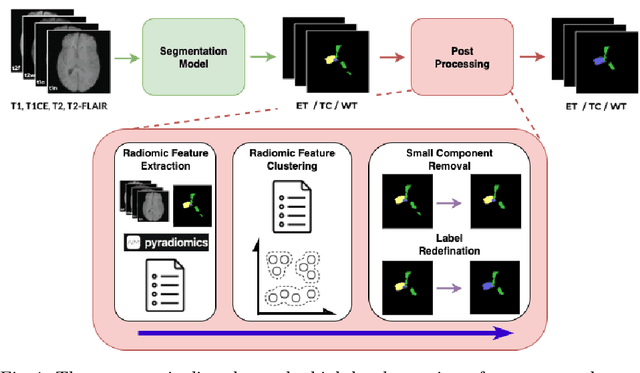
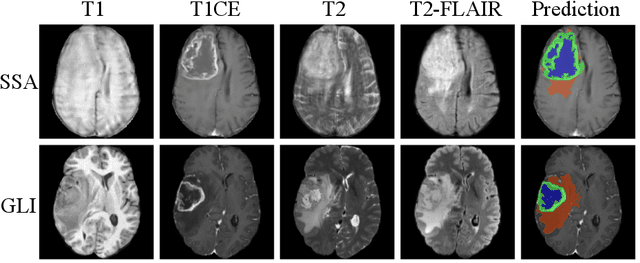

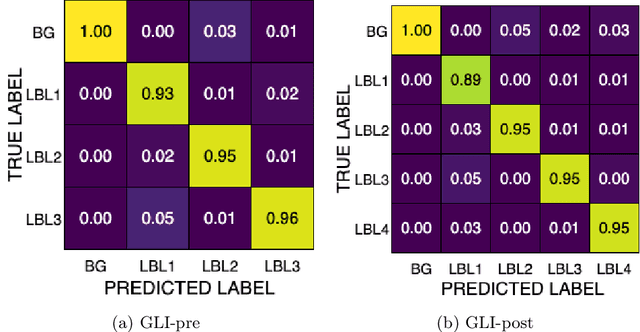
Abstract:Gliomas are the most common malignant brain tumors in adults and are among the most lethal. Despite aggressive treatment, the median survival rate is less than 15 months. Accurate multiparametric MRI (mpMRI) tumor segmentation is critical for surgical planning, radiotherapy, and disease monitoring. While deep learning models have improved the accuracy of automated segmentation, large-scale pre-trained models generalize poorly and often underperform, producing systematic errors such as false positives, label swaps, and slice discontinuities in slices. These limitations are further compounded by unequal access to GPU resources and the growing environmental cost of large-scale model training. In this work, we propose adaptive post-processing techniques to refine the quality of glioma segmentations produced by large-scale pretrained models developed for various types of tumors. We demonstrated the techniques in multiple BraTS 2025 segmentation challenge tasks, with the ranking metric improving by 14.9 % for the sub-Saharan Africa challenge and 0.9% for the adult glioma challenge. This approach promotes a shift in brain tumor segmentation research from increasingly complex model architectures to efficient, clinically aligned post-processing strategies that are precise, computationally fair, and sustainable.
Magnetic Resonance Imaging Feature-Based Subtyping and Model Ensemble for Enhanced Brain Tumor Segmentation
Dec 05, 2024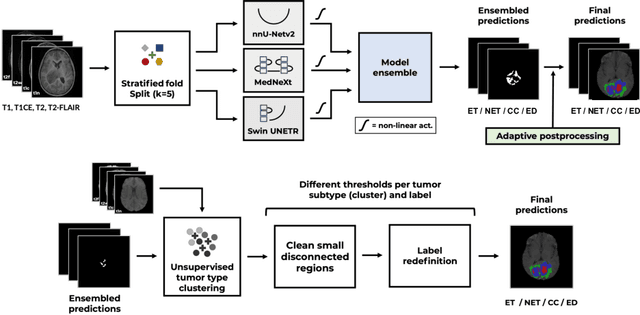
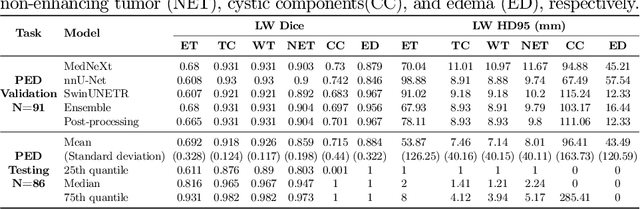
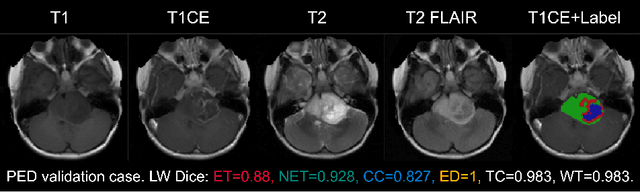
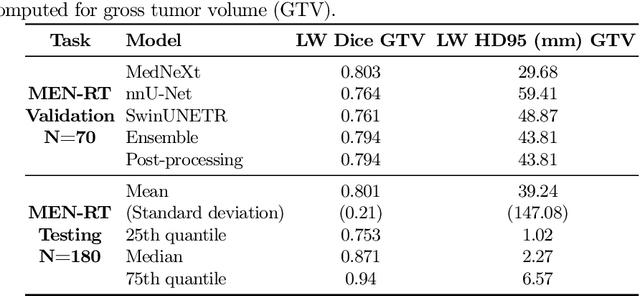
Abstract:Accurate and automatic segmentation of brain tumors in multi-parametric magnetic resonance imaging (mpMRI) is essential for quantitative measurements, which play an increasingly important role in clinical diagnosis and prognosis. The International Brain Tumor Segmentation (BraTS) Challenge 2024 offers a unique benchmarking opportunity, including various types of brain tumors in both adult and pediatric populations, such as pediatric brain tumors (PED), meningiomas (MEN-RT) and brain metastases (MET), among others. Compared to previous editions, BraTS 2024 has implemented changes to substantially increase clinical relevance, such as refined tumor regions for evaluation. We propose a deep learning-based ensemble approach that integrates state-of-the-art segmentation models. Additionally, we introduce innovative, adaptive pre- and post-processing techniques that employ MRI-based radiomic analyses to differentiate tumor subtypes. Given the heterogeneous nature of the tumors present in the BraTS datasets, this approach enhances the precision and generalizability of segmentation models. On the final testing sets, our method achieved mean lesion-wise Dice similarity coefficients of 0.926, 0.801, and 0.688 for the whole tumor in PED, MEN-RT, and MET, respectively. These results demonstrate the effectiveness of our approach in improving segmentation performance and generalizability for various brain tumor types.
Adult Glioma Segmentation in Sub-Saharan Africa using Transfer Learning on Stratified Finetuning Data
Dec 05, 2024



Abstract:Gliomas, a kind of brain tumor characterized by high mortality, present substantial diagnostic challenges in low- and middle-income countries, particularly in Sub-Saharan Africa. This paper introduces a novel approach to glioma segmentation using transfer learning to address challenges in resource-limited regions with minimal and low-quality MRI data. We leverage pre-trained deep learning models, nnU-Net and MedNeXt, and apply a stratified fine-tuning strategy using the BraTS2023-Adult-Glioma and BraTS-Africa datasets. Our method exploits radiomic analysis to create stratified training folds, model training on a large brain tumor dataset, and transfer learning to the Sub-Saharan context. A weighted model ensembling strategy and adaptive post-processing are employed to enhance segmentation accuracy. The evaluation of our proposed method on unseen validation cases on the BraTS-Africa 2024 task resulted in lesion-wise mean Dice scores of 0.870, 0.865, and 0.926, for enhancing tumor, tumor core, and whole tumor regions and was ranked first for the challenge. Our approach highlights the ability of integrated machine-learning techniques to bridge the gap between the medical imaging capabilities of resource-limited countries and established developed regions. By tailoring our methods to a target population's specific needs and constraints, we aim to enhance diagnostic capabilities in isolated environments. Our findings underscore the importance of approaches like local data integration and stratification refinement to address healthcare disparities, ensure practical applicability, and enhance impact.
Model Ensemble for Brain Tumor Segmentation in Magnetic Resonance Imaging
Sep 12, 2024



Abstract:Segmenting brain tumors in multi-parametric magnetic resonance imaging enables performing quantitative analysis in support of clinical trials and personalized patient care. This analysis provides the potential to impact clinical decision-making processes, including diagnosis and prognosis. In 2023, the well-established Brain Tumor Segmentation (BraTS) challenge presented a substantial expansion with eight tasks and 4,500 brain tumor cases. In this paper, we present a deep learning-based ensemble strategy that is evaluated for newly included tumor cases in three tasks: pediatric brain tumors (PED), intracranial meningioma (MEN), and brain metastases (MET). In particular, we ensemble outputs from state-of-the-art nnU-Net and Swin UNETR models on a region-wise basis. Furthermore, we implemented a targeted post-processing strategy based on a cross-validated threshold search to improve the segmentation results for tumor sub-regions. The evaluation of our proposed method on unseen test cases for the three tasks resulted in lesion-wise Dice scores for PED: 0.653, 0.809, 0.826; MEN: 0.876, 0.867, 0.849; and MET: 0.555, 0.6, 0.58; for the enhancing tumor, tumor core, and whole tumor, respectively. Our method was ranked first for PED, third for MEN, and fourth for MET, respectively.
Data Alchemy: Mitigating Cross-Site Model Variability Through Test Time Data Calibration
Jul 18, 2024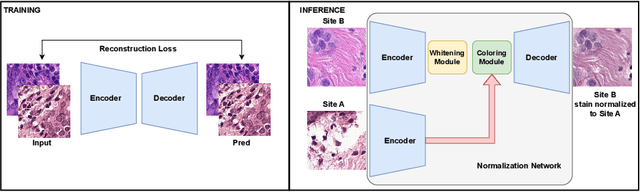
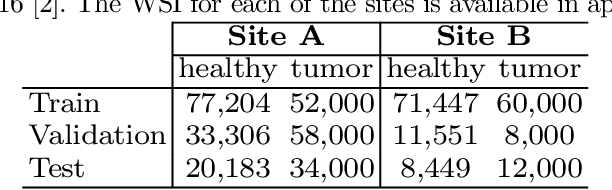
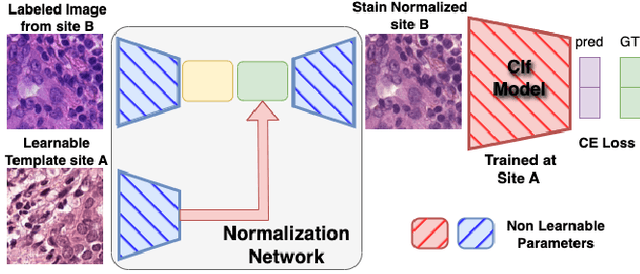
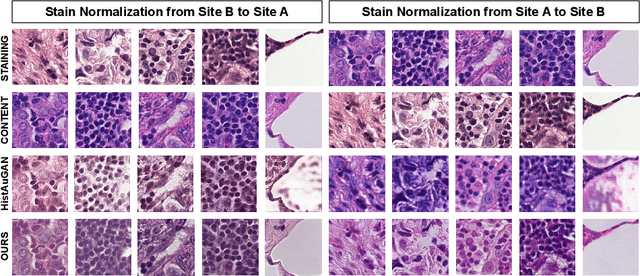
Abstract:Deploying deep learning-based imaging tools across various clinical sites poses significant challenges due to inherent domain shifts and regulatory hurdles associated with site-specific fine-tuning. For histopathology, stain normalization techniques can mitigate discrepancies, but they often fall short of eliminating inter-site variations. Therefore, we present Data Alchemy, an explainable stain normalization method combined with test time data calibration via a template learning framework to overcome barriers in cross-site analysis. Data Alchemy handles shifts inherent to multi-site data and minimizes them without needing to change the weights of the normalization or classifier networks. Our approach extends to unseen sites in various clinical settings where data domain discrepancies are unknown. Extensive experiments highlight the efficacy of our framework in tumor classification in hematoxylin and eosin-stained patches. Our explainable normalization method boosts classification tasks' area under the precision-recall curve(AUPR) by 0.165, 0.545 to 0.710. Additionally, Data Alchemy further reduces the multisite classification domain gap, by improving the 0.710 AUPR an additional 0.142, elevating classification performance further to 0.852, from 0.545. Our Data Alchemy framework can popularize precision medicine with minimal operational overhead by allowing for the seamless integration of pre-trained deep learning-based clinical tools across multiple sites.
Harvesting Private Medical Images in Federated Learning Systems with Crafted Models
Jul 13, 2024Abstract:Federated learning (FL) allows a set of clients to collaboratively train a machine-learning model without exposing local training samples. In this context, it is considered to be privacy-preserving and hence has been adopted by medical centers to train machine-learning models over private data. However, in this paper, we propose a novel attack named MediLeak that enables a malicious parameter server to recover high-fidelity patient images from the model updates uploaded by the clients. MediLeak requires the server to generate an adversarial model by adding a crafted module in front of the original model architecture. It is published to the clients in the regular FL training process and each client conducts local training on it to generate corresponding model updates. Then, based on the FL protocol, the model updates are sent back to the server and our proposed analytical method recovers private data from the parameter updates of the crafted module. We provide a comprehensive analysis for MediLeak and show that it can successfully break the state-of-the-art cryptographic secure aggregation protocols, designed to protect the FL systems from privacy inference attacks. We implement MediLeak on the MedMNIST and COVIDx CXR-4 datasets. The results show that MediLeak can nearly perfectly recover private images with high recovery rates and quantitative scores. We further perform downstream tasks such as disease classification with the recovered data, where our results show no significant performance degradation compared to using the original training samples.
Lung-CADex: Fully automatic Zero-Shot Detection and Classification of Lung Nodules in Thoracic CT Images
Jul 02, 2024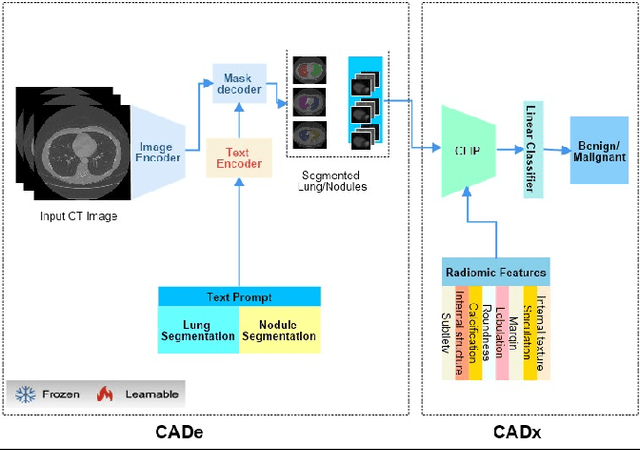
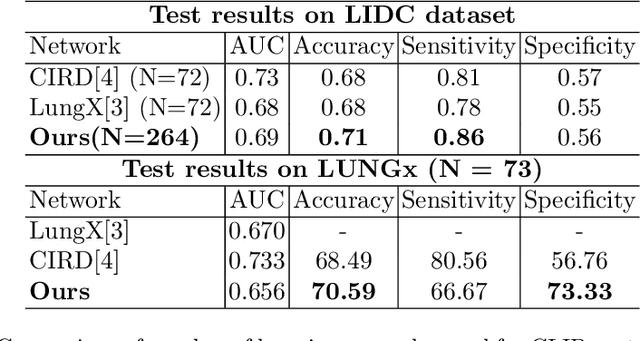
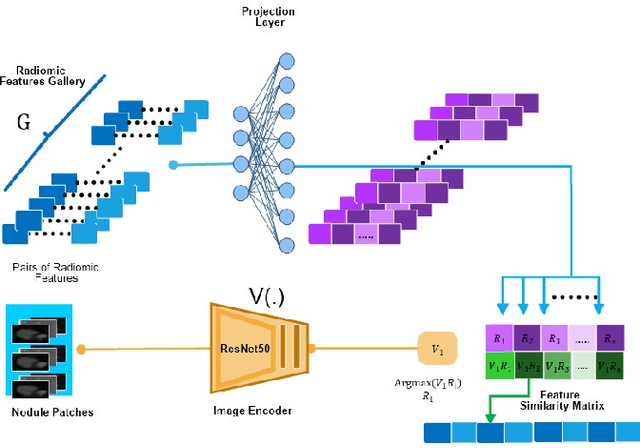
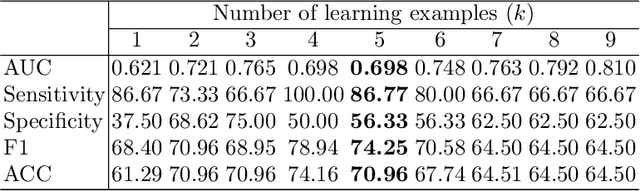
Abstract:Lung cancer has been one of the major threats to human life for decades. Computer-aided diagnosis can help with early lung nodul detection and facilitate subsequent nodule characterization. Large Visual Language models (VLMs) have been found effective for multiple downstream medical tasks that rely on both imaging and text data. However, lesion level detection and subsequent diagnosis using VLMs have not been explored yet. We propose CADe, for segmenting lung nodules in a zero-shot manner using a variant of the Segment Anything Model called MedSAM. CADe trains on a prompt suite on input computed tomography (CT) scans by using the CLIP text encoder through prefix tuning. We also propose, CADx, a method for the nodule characterization as benign/malignant by making a gallery of radiomic features and aligning image-feature pairs through contrastive learning. Training and validation of CADe and CADx have been done using one of the largest publicly available datasets, called LIDC. To check the generalization ability of the model, it is also evaluated on a challenging dataset, LUNGx. Our experimental results show that the proposed methods achieve a sensitivity of 0.86 compared to 0.76 that of other fully supervised methods.The source code, datasets and pre-processed data can be accessed using the link:
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge