Robert A. Avery
Quantitative Metrics for Benchmarking Medical Image Harmonization
Feb 06, 2024



Abstract:Image harmonization is an important preprocessing strategy to address domain shifts arising from data acquired using different machines and scanning protocols in medical imaging. However, benchmarking the effectiveness of harmonization techniques has been a challenge due to the lack of widely available standardized datasets with ground truths. In this context, we propose three metrics: two intensity harmonization metrics and one anatomy preservation metric for medical images during harmonization, where no ground truths are required. Through extensive studies on a dataset with available harmonization ground truth, we demonstrate that our metrics are correlated with established image quality assessment metrics. We show how these novel metrics may be applied to real-world scenarios where no harmonization ground truth exists. Additionally, we provide insights into different interpretations of the metric values, shedding light on their significance in the context of the harmonization process. As a result of our findings, we advocate for the adoption of these quantitative harmonization metrics as a standard for benchmarking the performance of image harmonization techniques.
Harmonization Across Imaging Locations(HAIL): One-Shot Learning for Brain MRI
Aug 21, 2023Abstract:For machine learning-based prognosis and diagnosis of rare diseases, such as pediatric brain tumors, it is necessary to gather medical imaging data from multiple clinical sites that may use different devices and protocols. Deep learning-driven harmonization of radiologic images relies on generative adversarial networks (GANs). However, GANs notoriously generate pseudo structures that do not exist in the original training data, a phenomenon known as "hallucination". To prevent hallucination in medical imaging, such as magnetic resonance images (MRI) of the brain, we propose a one-shot learning method where we utilize neural style transfer for harmonization. At test time, the method uses one image from a clinical site to generate an image that matches the intensity scale of the collaborating sites. Our approach combines learning a feature extractor, neural style transfer, and adaptive instance normalization. We further propose a novel strategy to evaluate the effectiveness of image harmonization approaches with evaluation metrics that both measure image style harmonization and assess the preservation of anatomical structures. Experimental results demonstrate the effectiveness of our method in preserving patient anatomy while adjusting the image intensities to a new clinical site. Our general harmonization model can be used on unseen data from new sites, making it a valuable tool for real-world medical applications and clinical trials.
Partitioned Shape Modeling with On-the-Fly Sparse Appearance Learning for Anterior Visual Pathway Segmentation
Aug 05, 2015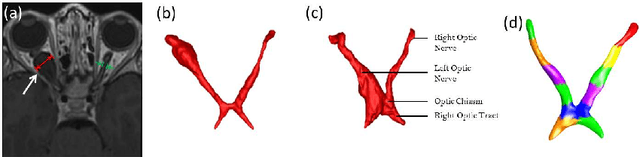
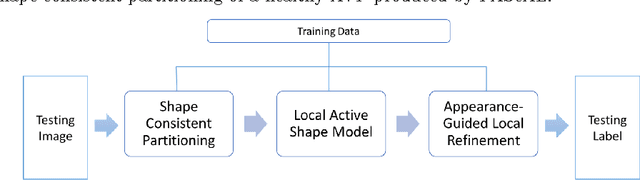
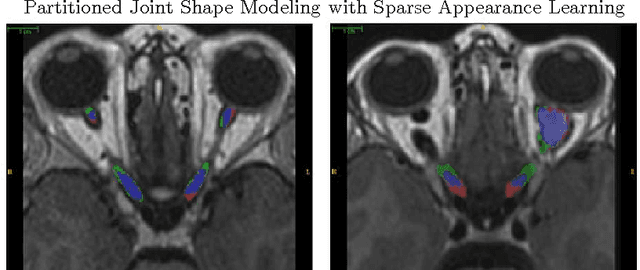
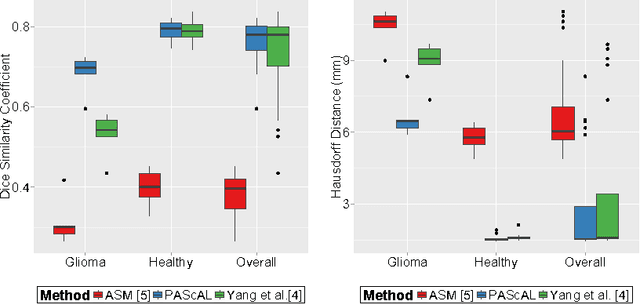
Abstract:MRI quantification of cranial nerves such as anterior visual pathway (AVP) in MRI is challenging due to their thin small size, structural variation along its path, and adjacent anatomic structures. Segmentation of pathologically abnormal optic nerve (e.g. optic nerve glioma) poses additional challenges due to changes in its shape at unpredictable locations. In this work, we propose a partitioned joint statistical shape model approach with sparse appearance learning for the segmentation of healthy and pathological AVP. Our main contributions are: (1) optimally partitioned statistical shape models for the AVP based on regional shape variations for greater local flexibility of statistical shape model; (2) refinement model to accommodate pathological regions as well as areas of subtle variation by training the model on-the-fly using the initial segmentation obtained in (1); (3) hierarchical deformable framework to incorporate scale information in partitioned shape and appearance models. Our method, entitled PAScAL (PArtitioned Shape and Appearance Learning), was evaluated on 21 MRI scans (15 healthy + 6 glioma cases) from pediatric patients (ages 2-17). The experimental results show that the proposed localized shape and sparse appearance-based learning approach significantly outperforms segmentation approaches in the analysis of pathological data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge