Juan J. Cerrolaza
Explainable Shape Analysis through Deep Hierarchical Generative Models: Application to Cardiac Remodeling
Jun 28, 2019
Abstract:Quantification of anatomical shape changes still relies on scalar global indexes which are largely insensitive to regional or asymmetric modifications. Accurate assessment of pathology-driven anatomical remodeling is a crucial step for the diagnosis and treatment of heart conditions. Deep learning approaches have recently achieved wide success in the analysis of medical images, but they lack interpretability in the feature extraction and decision processes. In this work, we propose a new interpretable deep learning model for shape analysis. In particular, we exploit deep generative networks to model a population of anatomical segmentations through a hierarchy of conditional latent variables. At the highest level of this hierarchy, a two-dimensional latent space is simultaneously optimised to discriminate distinct clinical conditions, enabling the direct visualisation of the classification space. Moreover, the anatomical variability encoded by this discriminative latent space can be visualised in the segmentation space thanks to the generative properties of the model, making the classification task transparent. This approach yielded high accuracy in the categorisation of healthy and remodelled hearts when tested on unseen segmentations from our own multi-centre dataset as well as in an external validation set. More importantly, it enabled the visualisation in three-dimensions of the most discriminative anatomical features between the two conditions. The proposed approach scales effectively to large populations, facilitating high-throughput analysis of normal anatomy and pathology in large-scale studies of volumetric imaging.
3D High-Resolution Cardiac Segmentation Reconstruction from 2D Views using Conditional Variational Autoencoders
Feb 28, 2019


Abstract:Accurate segmentation of heart structures imaged by cardiac MR is key for the quantitative analysis of pathology. High-resolution 3D MR sequences enable whole-heart structural imaging but are time-consuming, expensive to acquire and they often require long breath holds that are not suitable for patients. Consequently, multiplanar breath-hold 2D cine sequences are standard practice but are disadvantaged by lack of whole-heart coverage and low through-plane resolution. To address this, we propose a conditional variational autoencoder architecture able to learn a generative model of 3D high-resolution left ventricular (LV) segmentations which is conditioned on three 2D LV segmentations of one short-axis and two long-axis images. By only employing these three 2D segmentations, our model can efficiently reconstruct the 3D high-resolution LV segmentation of a subject. When evaluated on 400 unseen healthy volunteers, our model yielded an average Dice score of $87.92 \pm 0.15$ and outperformed competing architectures.
Computational Anatomy for Multi-Organ Analysis in Medical Imaging: A Review
Dec 20, 2018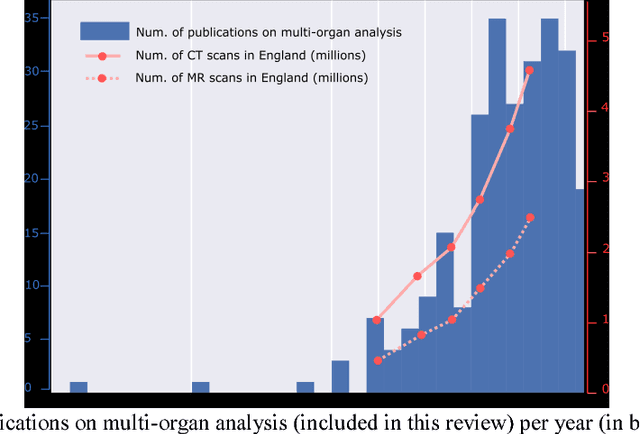
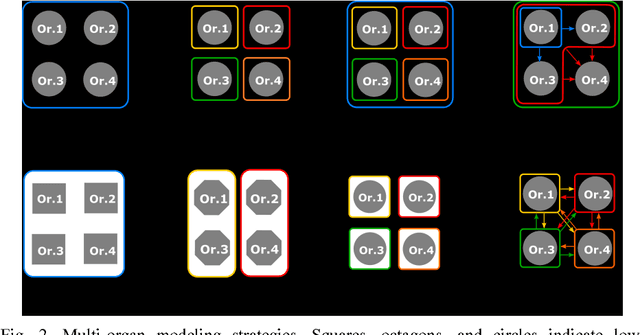
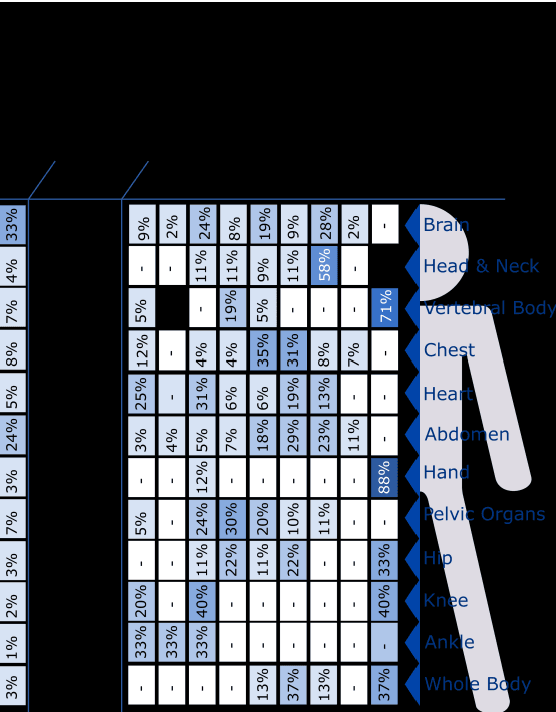
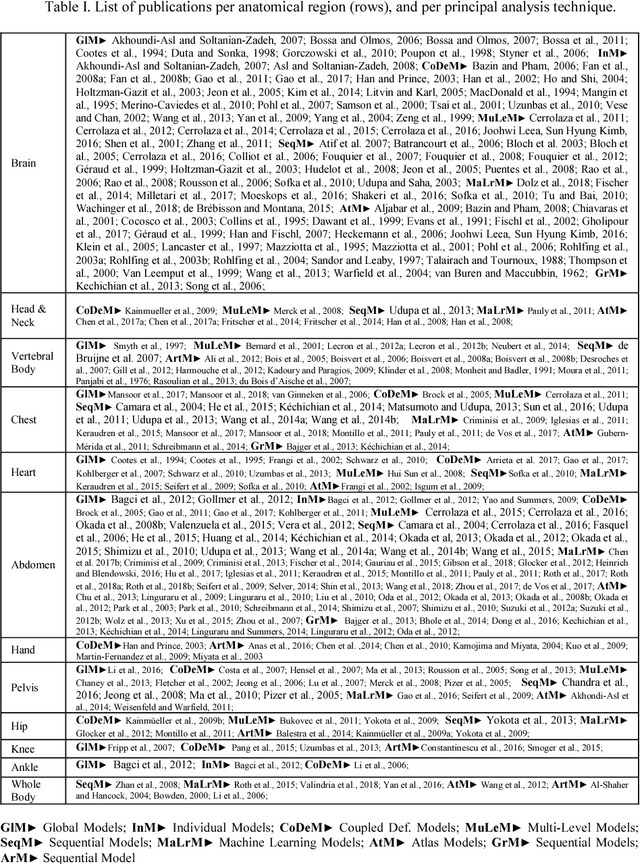
Abstract:The medical image analysis field has traditionally been focused on the development of organ-, and disease-specific methods. Recently, the interest in the development of more 20 comprehensive computational anatomical models has grown, leading to the creation of multi-organ models. Multi-organ approaches, unlike traditional organ-specific strategies, incorporate inter-organ relations into the model, thus leading to a more accurate representation of the complex human anatomy. Inter-organ relations are not only spatial, but also functional and physiological. Over the years, the strategies 25 proposed to efficiently model multi-organ structures have evolved from the simple global modeling, to more sophisticated approaches such as sequential, hierarchical, or machine learning-based models. In this paper, we present a review of the state of the art on multi-organ analysis and associated computation anatomy methodology. The manuscript follows a methodology-based classification of the different techniques 30 available for the analysis of multi-organs and multi-anatomical structures, from techniques using point distribution models to the most recent deep learning-based approaches. With more than 300 papers included in this review, we reflect on the trends and challenges of the field of computational anatomy, the particularities of each anatomical region, and the potential of multi-organ analysis to increase the impact of 35 medical imaging applications on the future of healthcare.
Standard Plane Detection in 3D Fetal Ultrasound Using an Iterative Transformation Network
Oct 07, 2018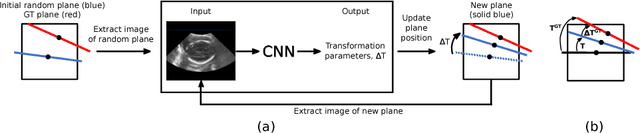
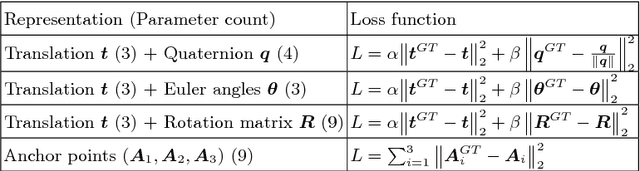
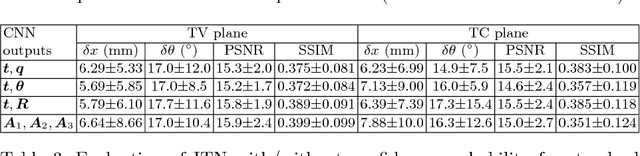
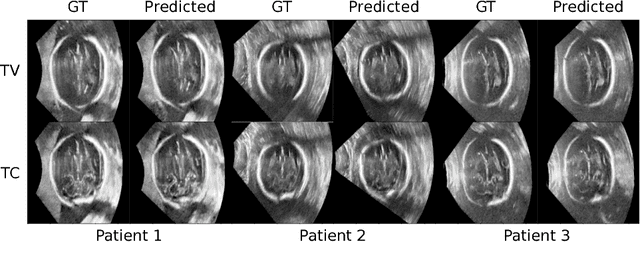
Abstract:Standard scan plane detection in fetal brain ultrasound (US) forms a crucial step in the assessment of fetal development. In clinical settings, this is done by manually manoeuvring a 2D probe to the desired scan plane. With the advent of 3D US, the entire fetal brain volume containing these standard planes can be easily acquired. However, manual standard plane identification in 3D volume is labour-intensive and requires expert knowledge of fetal anatomy. We propose a new Iterative Transformation Network (ITN) for the automatic detection of standard planes in 3D volumes. ITN uses a convolutional neural network to learn the relationship between a 2D plane image and the transformation parameters required to move that plane towards the location/orientation of the standard plane in the 3D volume. During inference, the current plane image is passed iteratively to the network until it converges to the standard plane location. We explore the effect of using different transformation representations as regression outputs of ITN. Under a multi-task learning framework, we introduce additional classification probability outputs to the network to act as confidence measures for the regressed transformation parameters in order to further improve the localisation accuracy. When evaluated on 72 US volumes of fetal brain, our method achieves an error of 3.83mm/12.7 degrees and 3.80mm/12.6 degrees for the transventricular and transcerebellar planes respectively and takes 0.46s per plane. Source code is publicly available at https://github.com/yuanwei1989/plane-detection.
* 8 pages, 2 figures, accepted for MICCAI 2018; Added link to source code
Fast Multiple Landmark Localisation Using a Patch-based Iterative Network
Oct 07, 2018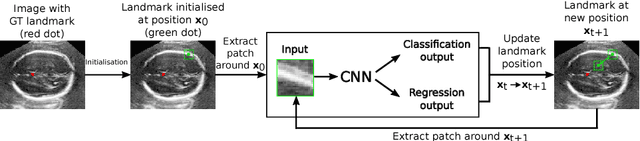
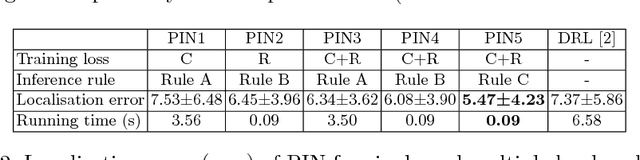
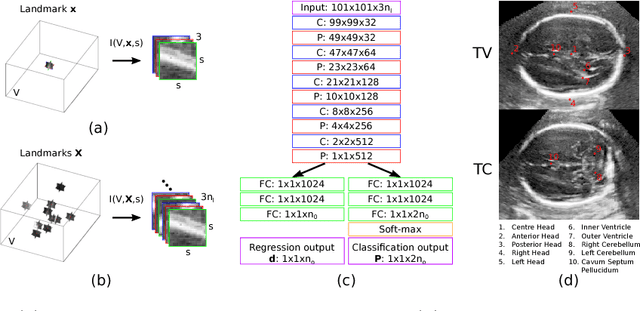
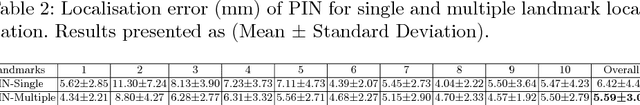
Abstract:We propose a new Patch-based Iterative Network (PIN) for fast and accurate landmark localisation in 3D medical volumes. PIN utilises a Convolutional Neural Network (CNN) to learn the spatial relationship between an image patch and anatomical landmark positions. During inference, patches are repeatedly passed to the CNN until the estimated landmark position converges to the true landmark location. PIN is computationally efficient since the inference stage only selectively samples a small number of patches in an iterative fashion rather than a dense sampling at every location in the volume. Our approach adopts a multi-task learning framework that combines regression and classification to improve localisation accuracy. We extend PIN to localise multiple landmarks by using principal component analysis, which models the global anatomical relationships between landmarks. We have evaluated PIN using 72 3D ultrasound images from fetal screening examinations. PIN achieves quantitatively an average landmark localisation error of 5.59mm and a runtime of 0.44s to predict 10 landmarks per volume. Qualitatively, anatomical 2D standard scan planes derived from the predicted landmark locations are visually similar to the clinical ground truth. Source code is publicly available at https://github.com/yuanwei1989/landmark-detection.
* 8 pages, 4 figures, Accepted for MICCAI 2018
A Generic Approach to Lung Field Segmentation from Chest Radiographs using Deep Space and Shape Learning
Jul 11, 2018



Abstract:Computer-aided diagnosis (CAD) techniques for lung field segmentation from chest radiographs (CXR) have been proposed for adult cohorts, but rarely for pediatric subjects. Statistical shape models (SSMs), the workhorse of most state-of-the-art CXR-based lung field segmentation methods, do not efficiently accommodate shape variation of the lung field during the pediatric developmental stages. The main contributions of our work are: (1) a generic lung field segmentation framework from CXR accommodating large shape variation for adult and pediatric cohorts; (2) a deep representation learning detection mechanism, \emph{ensemble space learning}, for robust object localization; and (3) \emph{marginal shape deep learning} for the shape deformation parameter estimation. Unlike the iterative approach of conventional SSMs, the proposed shape learning mechanism transforms the parameter space into marginal subspaces that are solvable efficiently using the recursive representation learning mechanism. Furthermore, our method is the first to include the challenging retro-cardiac region in the CXR-based lung segmentation for accurate lung capacity estimation. The framework is evaluated on 668 CXRs of patients between 3 month to 89 year of age. We obtain a mean Dice similarity coefficient of $0.96\pm0.03$ (including the retro-cardiac region). For a given accuracy, the proposed approach is also found to be faster than conventional SSM-based iterative segmentation methods. The computational simplicity of the proposed generic framework could be similarly applied to the fast segmentation of other deformable objects.
Partitioned Shape Modeling with On-the-Fly Sparse Appearance Learning for Anterior Visual Pathway Segmentation
Aug 05, 2015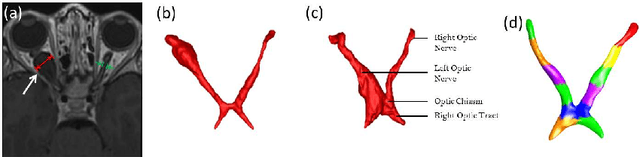
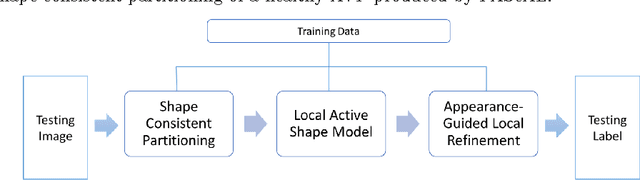
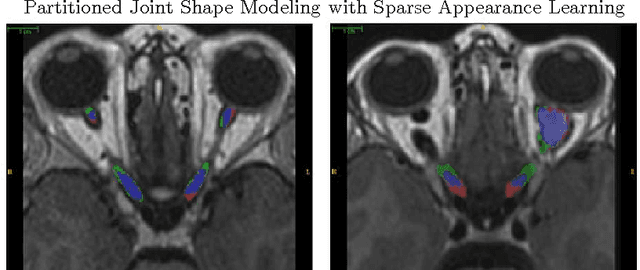
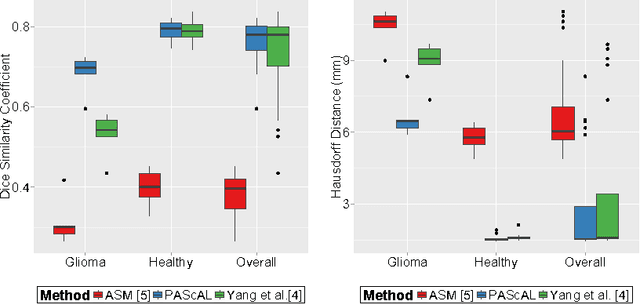
Abstract:MRI quantification of cranial nerves such as anterior visual pathway (AVP) in MRI is challenging due to their thin small size, structural variation along its path, and adjacent anatomic structures. Segmentation of pathologically abnormal optic nerve (e.g. optic nerve glioma) poses additional challenges due to changes in its shape at unpredictable locations. In this work, we propose a partitioned joint statistical shape model approach with sparse appearance learning for the segmentation of healthy and pathological AVP. Our main contributions are: (1) optimally partitioned statistical shape models for the AVP based on regional shape variations for greater local flexibility of statistical shape model; (2) refinement model to accommodate pathological regions as well as areas of subtle variation by training the model on-the-fly using the initial segmentation obtained in (1); (3) hierarchical deformable framework to incorporate scale information in partitioned shape and appearance models. Our method, entitled PAScAL (PArtitioned Shape and Appearance Learning), was evaluated on 21 MRI scans (15 healthy + 6 glioma cases) from pediatric patients (ages 2-17). The experimental results show that the proposed localized shape and sparse appearance-based learning approach significantly outperforms segmentation approaches in the analysis of pathological data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge