Giacomo Tarroni
Ensembled Cold-Diffusion Restorations for Unsupervised Anomaly Detection
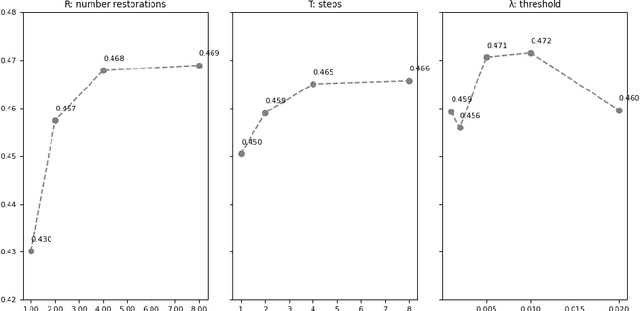
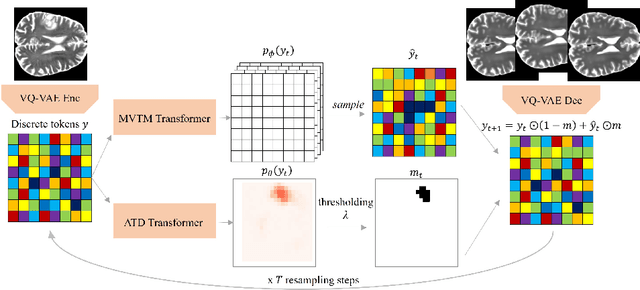
Jul 09, 2024Abstract:Unsupervised Anomaly Detection (UAD) methods aim to identify anomalies in test samples comparing them with a normative distribution learned from a dataset known to be anomaly-free. Approaches based on generative models offer interpretability by generating anomaly-free versions of test images, but are typically unable to identify subtle anomalies. Alternatively, approaches using feature modelling or self-supervised methods, such as the ones relying on synthetically generated anomalies, do not provide out-of-the-box interpretability. In this work, we present a novel method that combines the strengths of both strategies: a generative cold-diffusion pipeline (i.e., a diffusion-like pipeline which uses corruptions not based on noise) that is trained with the objective of turning synthetically-corrupted images back to their normal, original appearance. To support our pipeline we introduce a novel synthetic anomaly generation procedure, called DAG, and a novel anomaly score which ensembles restorations conditioned with different degrees of abnormality. Our method surpasses the prior state-of-the art for unsupervised anomaly detection in three different Brain MRI datasets.
Sample selection with noise rate estimation in noise learning of medical image analysis
Dec 23, 2023



Abstract:Deep learning techniques have demonstrated remarkable success in the field of medical image analysis. However, the existence of label noise within data significantly hampers its performance. In this paper, we introduce a novel noise-robust learning method which integrates noise rate estimation into sample selection approaches for handling noisy datasets. We first estimate the noise rate of a dataset with Linear Regression based on the distribution of loss values. Then, potentially noisy samples are excluded based on this estimated noise rate, and sparse regularization is further employed to improve the robustness of our deep learning model. Our proposed method is evaluated on five benchmark medical image classification datasets, including two datasets featuring 3D medical images. Experiments show that our method outperforms other existing noise-robust learning methods, especially when noise rate is very big.
DISYRE: Diffusion-Inspired SYnthetic REstoration for Unsupervised Anomaly Detection
Nov 26, 2023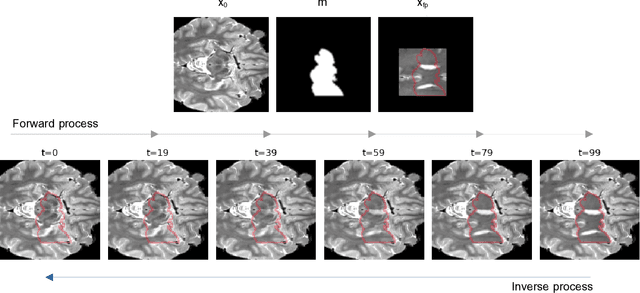
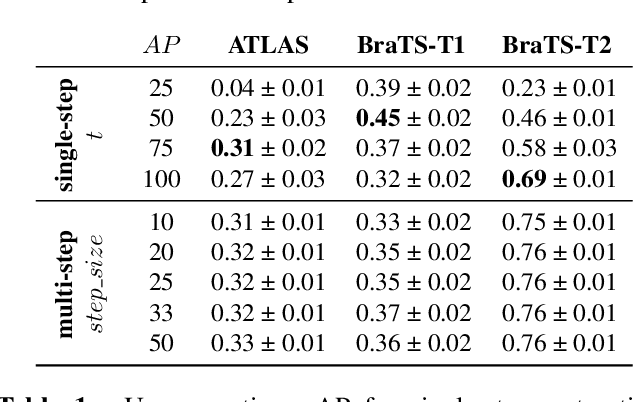
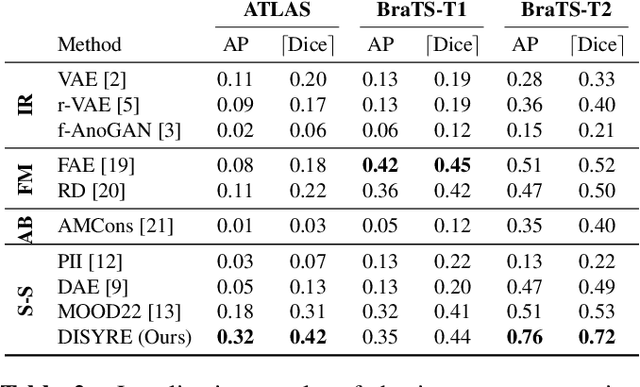
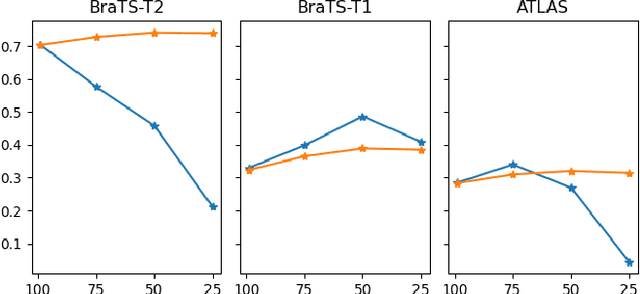
Abstract:Unsupervised Anomaly Detection (UAD) techniques aim to identify and localize anomalies without relying on annotations, only leveraging a model trained on a dataset known to be free of anomalies. Diffusion models learn to modify inputs $x$ to increase the probability of it belonging to a desired distribution, i.e., they model the score function $\nabla_x \log p(x)$. Such a score function is potentially relevant for UAD, since $\nabla_x \log p(x)$ is itself a pixel-wise anomaly score. However, diffusion models are trained to invert a corruption process based on Gaussian noise and the learned score function is unlikely to generalize to medical anomalies. This work addresses the problem of how to learn a score function relevant for UAD and proposes DISYRE: Diffusion-Inspired SYnthetic REstoration. We retain the diffusion-like pipeline but replace the Gaussian noise corruption with a gradual, synthetic anomaly corruption so the learned score function generalizes to medical, naturally occurring anomalies. We evaluate DISYRE on three common Brain MRI UAD benchmarks and substantially outperform other methods in two out of the three tasks.
ARIA: On the interaction between Architectures, Aggregation methods and Initializations in federated visual classification
Nov 24, 2023Abstract:Federated Learning (FL) is a collaborative training paradigm that allows for privacy-preserving learning of cross-institutional models by eliminating the exchange of sensitive data and instead relying on the exchange of model parameters between the clients and a server. Despite individual studies on how client models are aggregated, and, more recently, on the benefits of ImageNet pre-training, there is a lack of understanding of the effect the architecture chosen for the federation has, and of how the aforementioned elements interconnect. To this end, we conduct the first joint ARchitecture-Initialization-Aggregation study and benchmark ARIAs across a range of medical image classification tasks. We find that, contrary to current practices, ARIA elements have to be chosen together to achieve the best possible performance. Our results also shed light on good choices for each element depending on the task, the effect of normalisation layers, and the utility of SSL pre-training, pointing to potential directions for designing FL-specific architectures and training pipelines.
MIM-OOD: Generative Masked Image Modelling for Out-of-Distribution Detection in Medical Images
Aug 02, 2023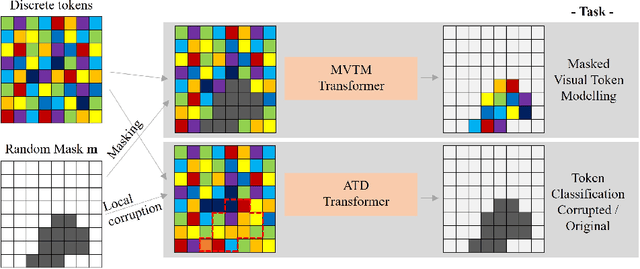

Abstract:Unsupervised Out-of-Distribution (OOD) detection consists in identifying anomalous regions in images leveraging only models trained on images of healthy anatomy. An established approach is to tokenize images and model the distribution of tokens with Auto-Regressive (AR) models. AR models are used to 1) identify anomalous tokens and 2) in-paint anomalous representations with in-distribution tokens. However, AR models are slow at inference time and prone to error accumulation issues which negatively affect OOD detection performance. Our novel method, MIM-OOD, overcomes both speed and error accumulation issues by replacing the AR model with two task-specific networks: 1) a transformer optimized to identify anomalous tokens and 2) a transformer optimized to in-paint anomalous tokens using masked image modelling (MIM). Our experiments with brain MRI anomalies show that MIM-OOD substantially outperforms AR models (DICE 0.458 vs 0.301) while achieving a nearly 25x speedup (9.5s vs 244s).
Harder synthetic anomalies to improve OoD detection in Medical Images
Aug 02, 2023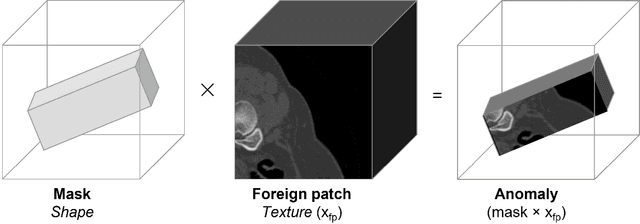
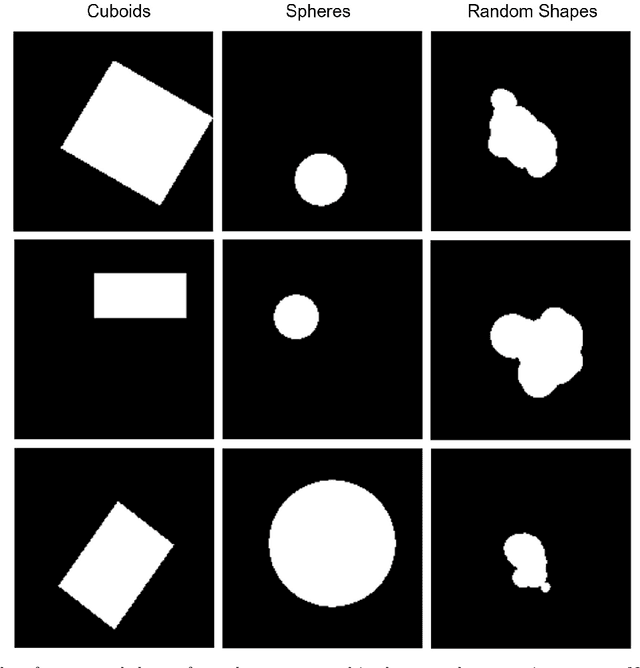
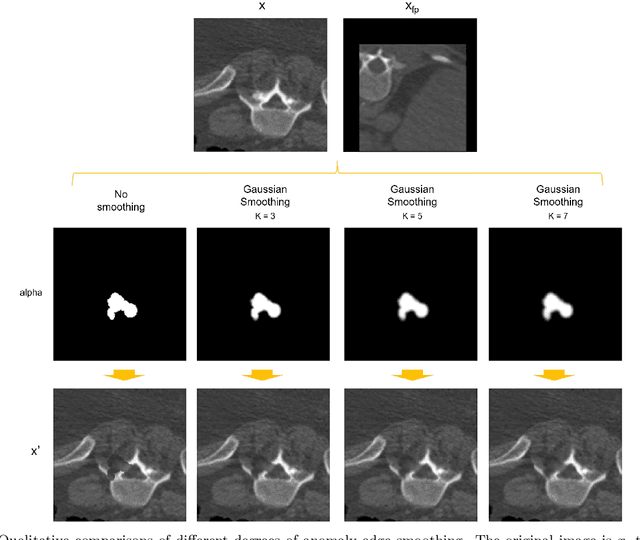
Abstract:Our method builds upon previous Medical Out-of-Distribution (MOOD) challenge winners that empirically show that synthetic local anomalies generated copying / interpolating foreign patches are useful to train segmentation networks able to generalize to unseen types of anomalies. In terms of the synthetic anomaly generation process, our contributions makes synthetic anomalies more heterogeneous and challenging by 1) using random shapes instead of squares and 2) smoothing the interpolation edge of anomalies so networks cannot rely on the high gradient between image - foreign patch to identify anomalies. Our experiments using the validation set of 2020 MOOD winners show that both contributions improved substantially the method performance. We used a standard 3D U-Net architecture as segmentation network, trained patch-wise in both brain and abdominal datasets. Our final challenge submission consisted of 10 U-Nets trained across 5 data folds with different configurations of the anomaly generation process. Our method achieved first position in both sample-wise and pixel-wise tasks in the 2022 edition of the Medical Out-of-Distribution held at MICCAI.
NeSy4VRD: A Multifaceted Resource for Neurosymbolic AI Research using Knowledge Graphs in Visual Relationship Detection
May 22, 2023Abstract:NeSy4VRD is a multifaceted resource designed to support the development of neurosymbolic AI (NeSy) research. NeSy4VRD re-establishes public access to the images of the VRD dataset and couples them with an extensively revised, quality-improved version of the VRD visual relationship annotations. Crucially, NeSy4VRD provides a well-aligned, companion OWL ontology that describes the dataset domain.It comes with open source infrastructure that provides comprehensive support for extensibility of the annotations (which, in turn, facilitates extensibility of the ontology), and open source code for loading the annotations to/from a knowledge graph. We are contributing NeSy4VRD to the computer vision, NeSy and Semantic Web communities to help foster more NeSy research using OWL-based knowledge graphs.
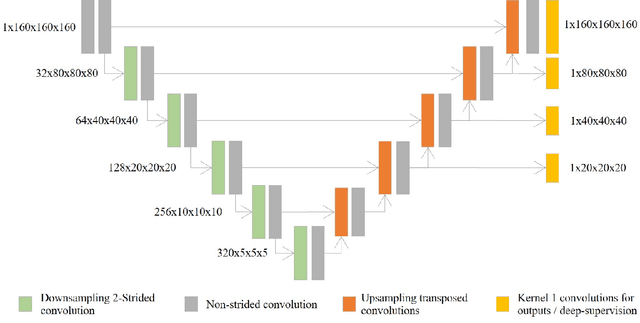
Implicit U-Net for volumetric medical image segmentation
Jun 30, 2022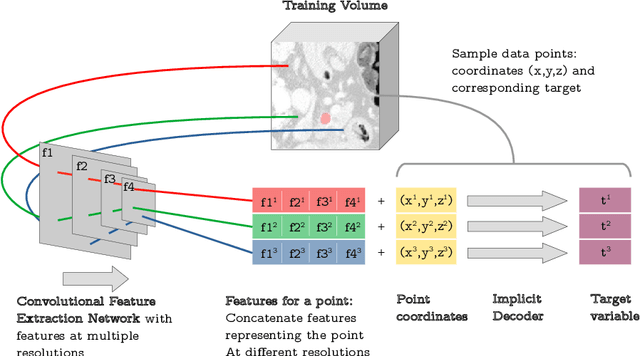
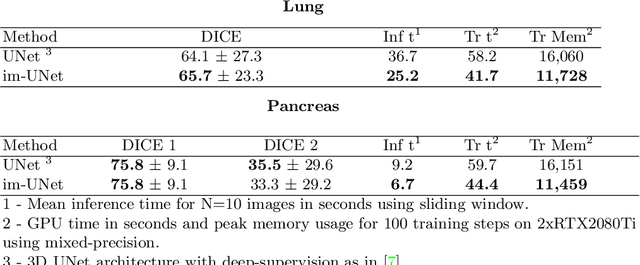
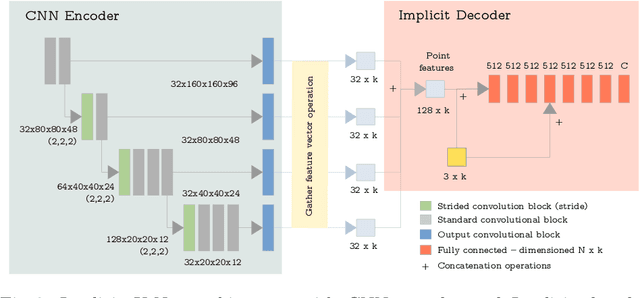
Abstract:U-Net has been the go-to architecture for medical image segmentation tasks, however computational challenges arise when extending the U-Net architecture to 3D images. We propose the Implicit U-Net architecture that adapts the efficient Implicit Representation paradigm to supervised image segmentation tasks. By combining a convolutional feature extractor with an implicit localization network, our implicit U-Net has 40% less parameters than the equivalent U-Net. Moreover, we propose training and inference procedures to capitalize sparse predictions. When comparing to an equivalent fully convolutional U-Net, Implicit U-Net reduces by approximately 30% inference and training time as well as training memory footprint while achieving comparable results in our experiments with two different abdominal CT scan datasets.
Enhancing MR Image Segmentation with Realistic Adversarial Data Augmentation
Aug 07, 2021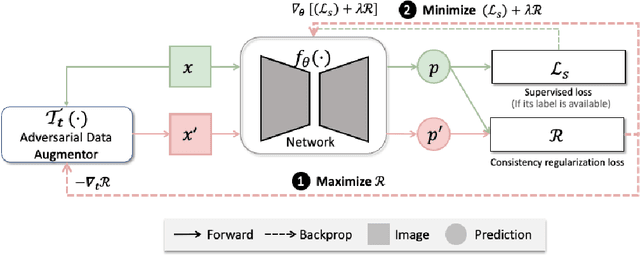
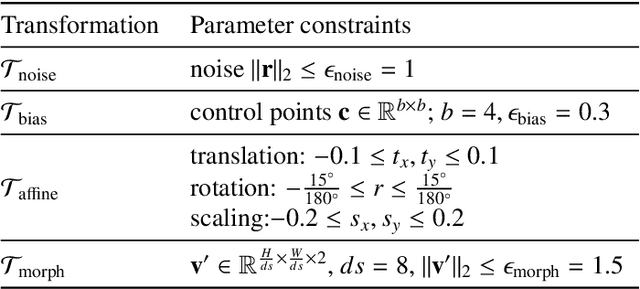
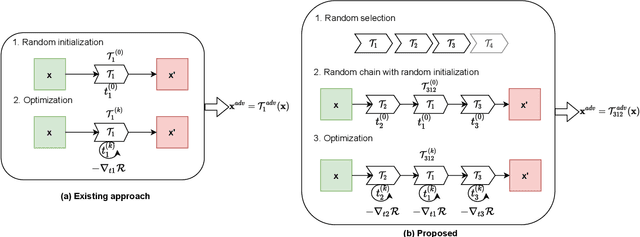
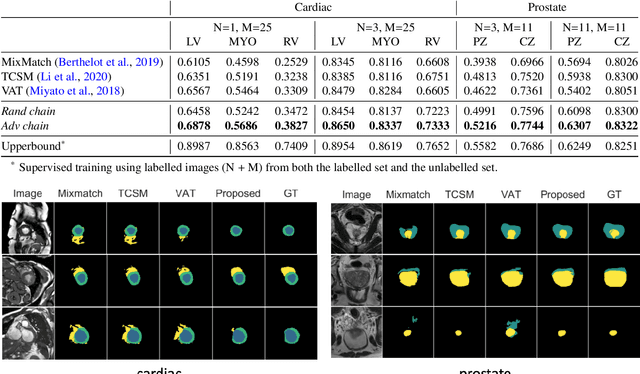
Abstract:The success of neural networks on medical image segmentation tasks typically relies on large labeled datasets for model training. However, acquiring and manually labeling a large medical image set is resource-intensive, expensive, and sometimes impractical due to data sharing and privacy issues. To address this challenge, we propose an adversarial data augmentation approach to improve the efficiency in utilizing training data and to enlarge the dataset via simulated but realistic transformations. Specifically, we present a generic task-driven learning framework, which jointly optimizes a data augmentation model and a segmentation network during training, generating informative examples to enhance network generalizability for the downstream task. The data augmentation model utilizes a set of photometric and geometric image transformations and chains them to simulate realistic complex imaging variations that could exist in magnetic resonance (MR) imaging. The proposed adversarial data augmentation does not rely on generative networks and can be used as a plug-in module in general segmentation networks. It is computationally efficient and applicable for both supervised and semi-supervised learning. We analyze and evaluate the method on two MR image segmentation tasks: cardiac segmentation and prostate segmentation. Results show that the proposed approach can alleviate the need for labeled data while improving model generalization ability, indicating its practical value in medical imaging applications.
Implicit field learning for unsupervised anomaly detection in medical images
Jun 09, 2021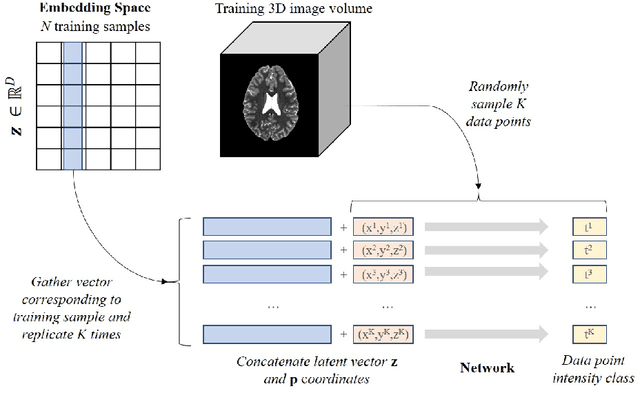
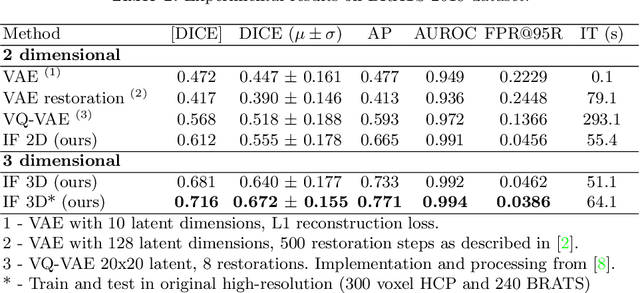
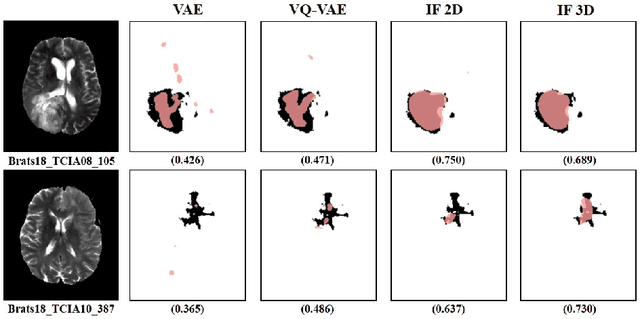
Abstract:We propose a novel unsupervised out-of-distribution detection method for medical images based on implicit fields image representations. In our approach, an auto-decoder feed-forward neural network learns the distribution of healthy images in the form of a mapping between spatial coordinates and probabilities over a proxy for tissue types. At inference time, the learnt distribution is used to retrieve, from a given test image, a restoration, i.e. an image maximally consistent with the input one but belonging to the healthy distribution. Anomalies are localized using the voxel-wise probability predicted by our model for the restored image. We tested our approach in the task of unsupervised localization of gliomas on brain MR images and compared it to several other VAE-based anomaly detection methods. Results show that the proposed technique substantially outperforms them (average DICE 0.640 vs 0.518 for the best performing VAE-based alternative) while also requiring considerably less computing time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge