Yuanwei Li
FADE: Few-shot/zero-shot Anomaly Detection Engine using Large Vision-Language Model
Aug 31, 2024



Abstract:Automatic image anomaly detection is important for quality inspection in the manufacturing industry. The usual unsupervised anomaly detection approach is to train a model for each object class using a dataset of normal samples. However, a more realistic problem is zero-/few-shot anomaly detection where zero or only a few normal samples are available. This makes the training of object-specific models challenging. Recently, large foundation vision-language models have shown strong zero-shot performance in various downstream tasks. While these models have learned complex relationships between vision and language, they are not specifically designed for the tasks of anomaly detection. In this paper, we propose the Few-shot/zero-shot Anomaly Detection Engine (FADE) which leverages the vision-language CLIP model and adjusts it for the purpose of industrial anomaly detection. Specifically, we improve language-guided anomaly segmentation 1) by adapting CLIP to extract multi-scale image patch embeddings that are better aligned with language and 2) by automatically generating an ensemble of text prompts related to industrial anomaly detection. 3) We use additional vision-based guidance from the query and reference images to further improve both zero-shot and few-shot anomaly detection. On the MVTec-AD (and VisA) dataset, FADE outperforms other state-of-the-art methods in anomaly segmentation with pixel-AUROC of 89.6% (91.5%) in zero-shot and 95.4% (97.5%) in 1-normal-shot. Code is available at https://github.com/BMVC-FADE/BMVC-FADE.
Standard Plane Detection in 3D Fetal Ultrasound Using an Iterative Transformation Network
Oct 07, 2018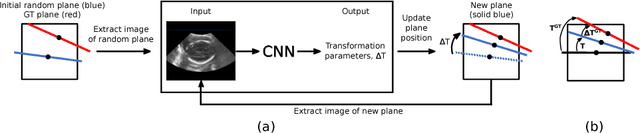
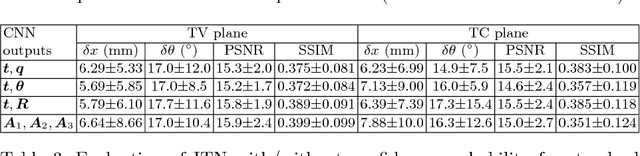
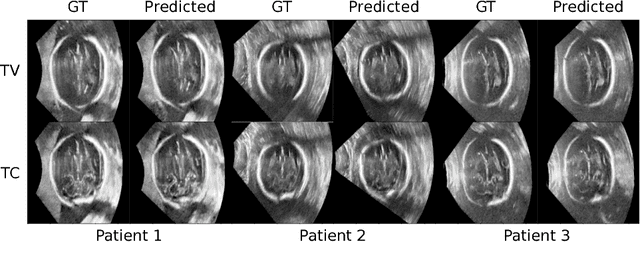
Abstract:Standard scan plane detection in fetal brain ultrasound (US) forms a crucial step in the assessment of fetal development. In clinical settings, this is done by manually manoeuvring a 2D probe to the desired scan plane. With the advent of 3D US, the entire fetal brain volume containing these standard planes can be easily acquired. However, manual standard plane identification in 3D volume is labour-intensive and requires expert knowledge of fetal anatomy. We propose a new Iterative Transformation Network (ITN) for the automatic detection of standard planes in 3D volumes. ITN uses a convolutional neural network to learn the relationship between a 2D plane image and the transformation parameters required to move that plane towards the location/orientation of the standard plane in the 3D volume. During inference, the current plane image is passed iteratively to the network until it converges to the standard plane location. We explore the effect of using different transformation representations as regression outputs of ITN. Under a multi-task learning framework, we introduce additional classification probability outputs to the network to act as confidence measures for the regressed transformation parameters in order to further improve the localisation accuracy. When evaluated on 72 US volumes of fetal brain, our method achieves an error of 3.83mm/12.7 degrees and 3.80mm/12.6 degrees for the transventricular and transcerebellar planes respectively and takes 0.46s per plane. Source code is publicly available at https://github.com/yuanwei1989/plane-detection.
* 8 pages, 2 figures, accepted for MICCAI 2018; Added link to source code
Fast Multiple Landmark Localisation Using a Patch-based Iterative Network
Oct 07, 2018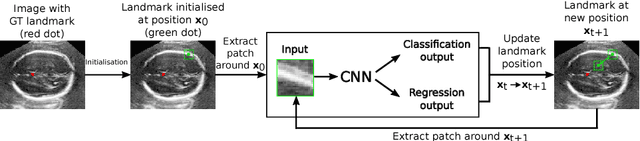
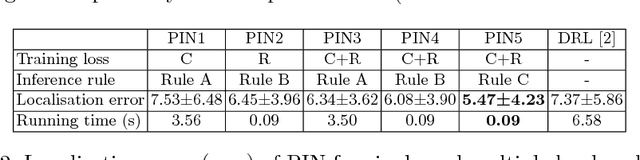
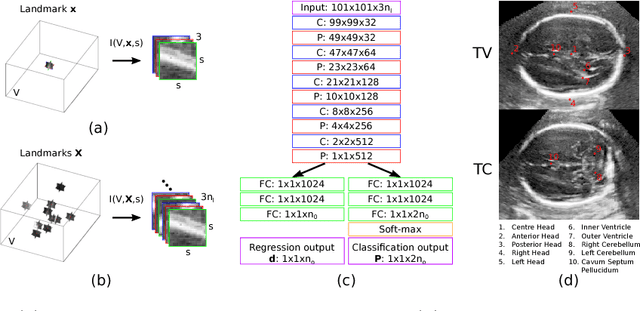
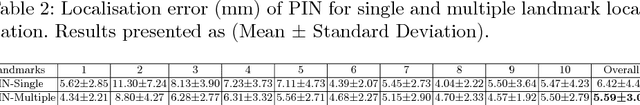
Abstract:We propose a new Patch-based Iterative Network (PIN) for fast and accurate landmark localisation in 3D medical volumes. PIN utilises a Convolutional Neural Network (CNN) to learn the spatial relationship between an image patch and anatomical landmark positions. During inference, patches are repeatedly passed to the CNN until the estimated landmark position converges to the true landmark location. PIN is computationally efficient since the inference stage only selectively samples a small number of patches in an iterative fashion rather than a dense sampling at every location in the volume. Our approach adopts a multi-task learning framework that combines regression and classification to improve localisation accuracy. We extend PIN to localise multiple landmarks by using principal component analysis, which models the global anatomical relationships between landmarks. We have evaluated PIN using 72 3D ultrasound images from fetal screening examinations. PIN achieves quantitatively an average landmark localisation error of 5.59mm and a runtime of 0.44s to predict 10 landmarks per volume. Qualitatively, anatomical 2D standard scan planes derived from the predicted landmark locations are visually similar to the clinical ground truth. Source code is publicly available at https://github.com/yuanwei1989/landmark-detection.
* 8 pages, 4 figures, Accepted for MICCAI 2018
Fully Automatic Myocardial Segmentation of Contrast Echocardiography Sequence Using Random Forests Guided by Shape Model
Jun 19, 2018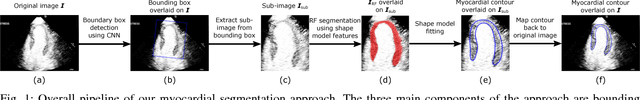
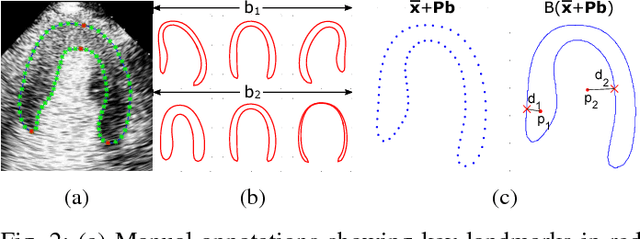
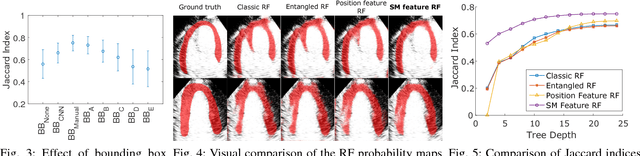
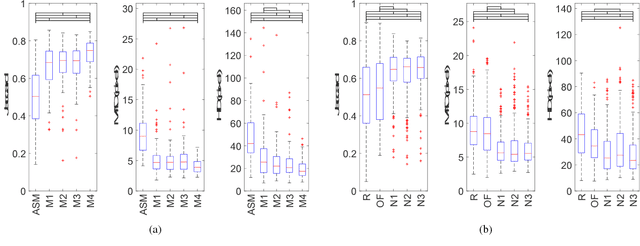
Abstract:Myocardial contrast echocardiography (MCE) is an imaging technique that assesses left ventricle function and myocardial perfusion for the detection of coronary artery diseases. Automatic MCE perfusion quantification is challenging and requires accurate segmentation of the myocardium from noisy and time-varying images. Random forests (RF) have been successfully applied to many medical image segmentation tasks. However, the pixel-wise RF classifier ignores contextual relationships between label outputs of individual pixels. RF which only utilizes local appearance features is also susceptible to data suffering from large intensity variations. In this paper, we demonstrate how to overcome the above limitations of classic RF by presenting a fully automatic segmentation pipeline for myocardial segmentation in full-cycle 2D MCE data. Specifically, a statistical shape model is used to provide shape prior information that guide the RF segmentation in two ways. First, a novel shape model (SM) feature is incorporated into the RF framework to generate a more accurate RF probability map. Second, the shape model is fitted to the RF probability map to refine and constrain the final segmentation to plausible myocardial shapes. We further improve the performance by introducing a bounding box detection algorithm as a preprocessing step in the segmentation pipeline. Our approach on 2D image is further extended to 2D+t sequence which ensures temporal consistency in the resultant sequence segmentations. When evaluated on clinical MCE data, our proposed method achieves notable improvement in segmentation accuracy and outperforms other state-of-the-art methods including the classic RF and its variants, active shape model and image registration.
Myocardial Segmentation of Contrast Echocardiograms Using Random Forests Guided by Shape Model
Jun 19, 2018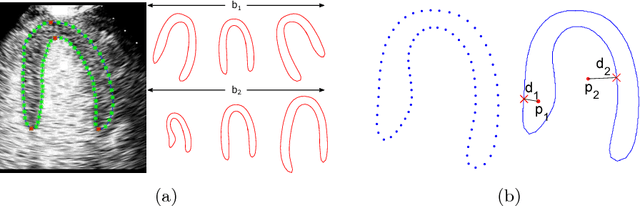

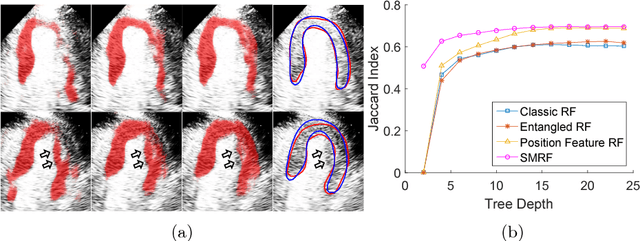
Abstract:Myocardial Contrast Echocardiography (MCE) with micro-bubble contrast agent enables myocardial perfusion quantification which is invaluable for the early detection of coronary artery diseases. In this paper, we proposed a new segmentation method called Shape Model guided Random Forests (SMRF) for the analysis of MCE data. The proposed method utilizes a statistical shape model of the myocardium to guide the Random Forest (RF) segmentation in two ways. First, we introduce a novel Shape Model (SM) feature which captures the global structure and shape of the myocardium to produce a more accurate RF probability map. Second, the shape model is fitted to the RF probability map to further refine and constrain the final segmentation to plausible myocardial shapes. Evaluated on clinical MCE images from 15 patients, our method obtained promising results (Dice=0.81, Jaccard=0.70, MAD=1.68 mm, HD=6.53 mm) and showed a notable improvement in segmentation accuracy over the classic RF and its variants.
Automatic View Planning with Multi-scale Deep Reinforcement Learning Agents
Jun 08, 2018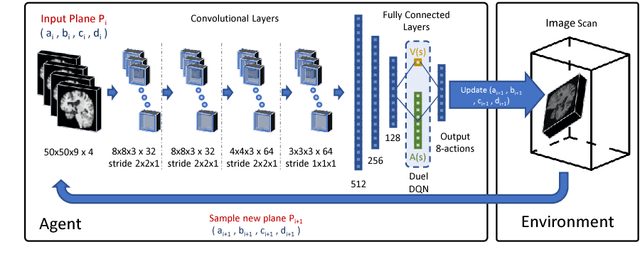
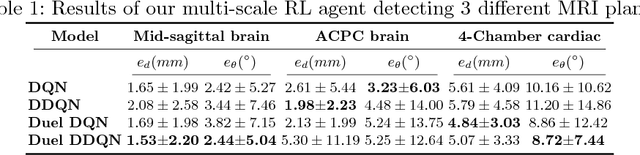
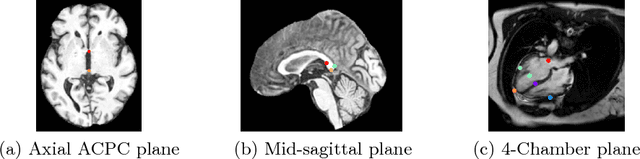
Abstract:We propose a fully automatic method to find standardized view planes in 3D image acquisitions. Standard view images are important in clinical practice as they provide a means to perform biometric measurements from similar anatomical regions. These views are often constrained to the native orientation of a 3D image acquisition. Navigating through target anatomy to find the required view plane is tedious and operator-dependent. For this task, we employ a multi-scale reinforcement learning (RL) agent framework and extensively evaluate several Deep Q-Network (DQN) based strategies. RL enables a natural learning paradigm by interaction with the environment, which can be used to mimic experienced operators. We evaluate our results using the distance between the anatomical landmarks and detected planes, and the angles between their normal vector and target. The proposed algorithm is assessed on the mid-sagittal and anterior-posterior commissure planes of brain MRI, and the 4-chamber long-axis plane commonly used in cardiac MRI, achieving accuracy of 1.53mm, 1.98mm and 4.84mm, respectively.
Human-level Performance On Automatic Head Biometrics In Fetal Ultrasound Using Fully Convolutional Neural Networks
Apr 24, 2018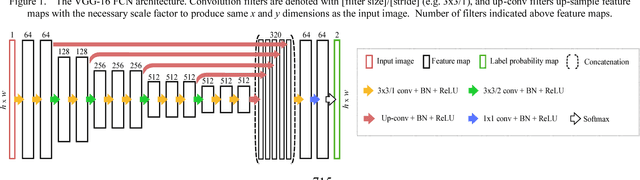

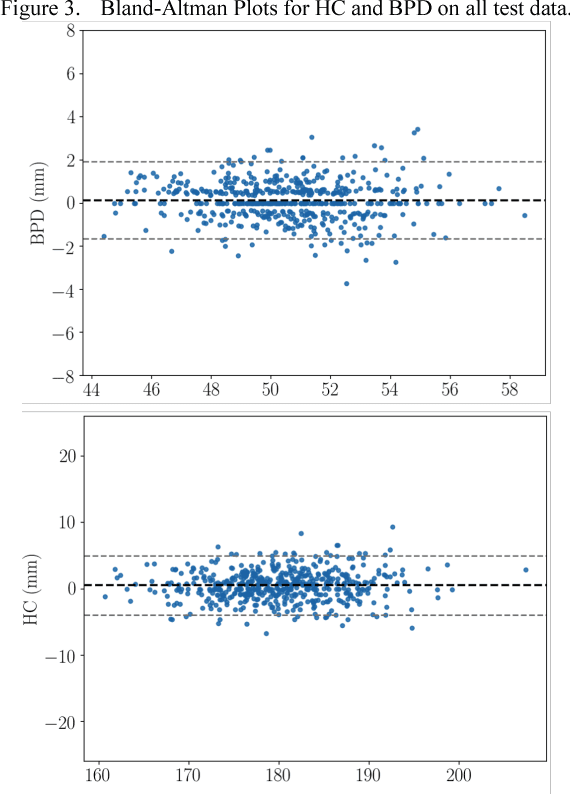
Abstract:Measurement of head biometrics from fetal ultrasonography images is of key importance in monitoring the healthy development of fetuses. However, the accurate measurement of relevant anatomical structures is subject to large inter-observer variability in the clinic. To address this issue, an automated method utilizing Fully Convolutional Networks (FCN) is proposed to determine measurements of fetal head circumference (HC) and biparietal diameter (BPD). An FCN was trained on approximately 2000 2D ultrasound images of the head with annotations provided by 45 different sonographers during routine screening examinations to perform semantic segmentation of the head. An ellipse is fitted to the resulting segmentation contours to mimic the annotation typically produced by a sonographer. The model's performance was compared with inter-observer variability, where two experts manually annotated 100 test images. Mean absolute model-expert error was slightly better than inter-observer error for HC (1.99mm vs 2.16mm), and comparable for BPD (0.61mm vs 0.59mm), as well as Dice coefficient (0.980 vs 0.980). Our results demonstrate that the model performs at a level similar to a human expert, and learns to produce accurate predictions from a large dataset annotated by many sonographers. Additionally, measurements are generated in near real-time at 15fps on a GPU, which could speed up clinical workflow for both skilled and trainee sonographers.
Pyramidal Gradient Matching for Optical Flow Estimation
Apr 11, 2017



Abstract:Initializing optical flow field by either sparse descriptor matching or dense patch matches has been proved to be particularly useful for capturing large displacements. In this paper, we present a pyramidal gradient matching approach that can provide dense matches for highly accurate and efficient optical flow estimation. A novel contribution of our method is that image gradient is used to describe image patches and proved to be able to produce robust matching. Therefore, our method is more efficient than methods that adopt special features (like SIFT) or patch distance metric. Moreover, we find that image gradient is scalable for optical flow estimation, which means we can use different levels of gradient feature (for example, full gradients or only direction information of gradients) to obtain different complexity without dramatic changes in accuracy. Another contribution is that we uncover the secrets of limited PatchMatch through a thorough analysis and design a pyramidal matching framework based these secrets. Our pyramidal matching framework is aimed at robust gradient matching and effective to grow inliers and reject outliers. In this framework, we present some special enhancements for outlier filtering in gradient matching. By initializing EpicFlow with our matches, experimental results show that our method is efficient and robust (ranking 1st on both clean pass and final pass of MPI Sintel dataset among published methods).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge