Meng-Xing Tang
Determining the utility of ultrafast nonlinear contrast enhanced and super resolution ultrasound for imaging microcirculation in the human small intestine
May 16, 2025Abstract:The regulation of intestinal blood flow is critical to gastrointestinal function. Imaging the intestinal mucosal micro-circulation in vivo has the potential to provide new insight into the gut physiology and pathophysiology. We aimed to determine whether ultrafast contrast enhanced ultrasound (CEUS) and super-resolution ultrasound localisation microscopy (SRUS/ULM) could be a useful tool for imaging the small intestine microcirculation in vivo non-invasively and for detecting changes in blood flow in the duodenum. Ultrafast CEUS and SRUS/ULM were used to image the small intestinal microcirculation in a cohort of 20 healthy volunteers (BMI<25). Participants were imaged while conscious and either having been fasted, or following ingestion of a liquid meal or water control, or under acute stress. For the first time we have performed ultrafast CEUS and ULM on the human small intestine, providing unprecedented resolution images of the intestinal microcirculation. We evaluated flow speed inside small vessels in healthy volunteers (2.78 +/- 0.05 mm/s, mean +/- SEM) and quantified changes in the perfusion of this microcirculation in response to nutrient ingestion. Perfusion of the microvasculature of the intestinal mucosa significantly increased post-prandially (36.2% +/- 12.2%, mean +/- SEM, p<0.05). The feasibility of 3D SRUS/ULM was also demonstrated. This study demonstrates the potential utility of ultrafast CEUS for assessing perfusion and detecting changes in blood flow in the duodenum. SRUS/ULM also proved a useful tool to image the microvascular blood flow in vivo non-invasively and to evaluate blood speed inside the microvasculature of the human small intestine.
Enhanced Acoustic Beamforming with Sub-Aperture Angular Multiply and Sum -- in vivo and in Human Demonstration
Jan 10, 2025



Abstract:Power Doppler ultrasound is in widespread clinical use for non-invasive vascular imaging but the most common current method - Delay and Sum (DAS) beamforming - suffers from limited resolution and high side-lobes. Here we propose the Sub-Aperture Angular Multiply and Sum (SAMAS) algorithm; it combines the advantages of two recent non-linear beamformers, Frame Multiply and Sum (FMAS) which uses signal temporal coherence and the acoustic sub-aperture (ASAP) algorithm, which uses signal spatial coherence, respectively. Following in vitro experiments to optimise the algorithm, particularly the use of phase information and sub-aperture pairing, it was evaluated in vivo, first in a rabbit kidney and then in human lymph node, using ultrafast ultrasound images obtained with intravenous contrast agents. The SAMAS algorithm improved the CNR and SNR across all tests, on average raising the CNR by 11 dB and the SNR by 18 dB over DAS in vivo. This work demonstrates a promising vascular imaging method that could have widespread clinical utility.
Online 4D Ultrasound-Guided Robotic Tracking Enables 3D Ultrasound Localisation Microscopy with Large Tissue Displacements
Sep 17, 2024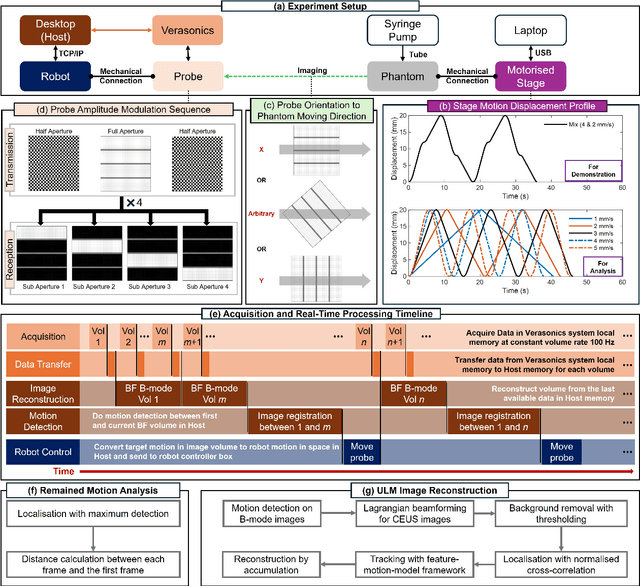
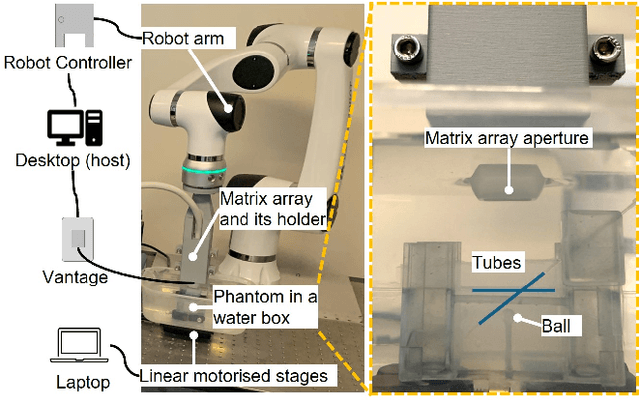
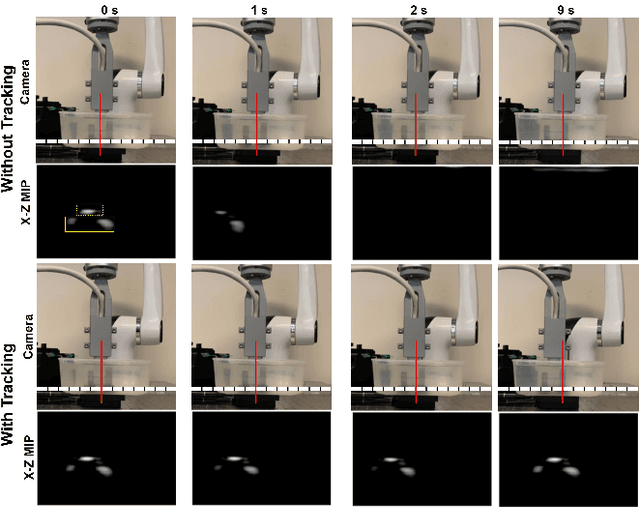
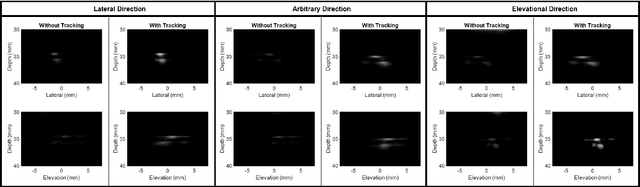
Abstract:Super-Resolution Ultrasound (SRUS) imaging through localising and tracking microbubbles, also known as Ultrasound Localisation Microscopy (ULM), has demonstrated significant potential for reconstructing microvasculature and flows with sub-diffraction resolution in clinical diagnostics. However, imaging organs with large tissue movements, such as those caused by respiration, presents substantial challenges. Existing methods often require breath holding to maintain accumulation accuracy, which limits data acquisition time and ULM image saturation. To improve image quality in the presence of large tissue movements, this study introduces an approach integrating high-frame-rate ultrasound with online precise robotic probe control. Tested on a microvasculature phantom with translation motions up to 20 mm, twice the aperture size of the matrix array used, our method achieved real-time tracking of the moving phantom and imaging volume rate at 85 Hz, keeping majority of the target volume in the imaging field of view. ULM images of the moving cross channels in the phantom were successfully reconstructed in post-processing, demonstrating the feasibility of super-resolution imaging under large tissue motions. This represents a significant step towards ULM imaging of organs with large motion.
Enhancing super-resolution ultrasound localisation through multi-frame deconvolution exploiting spatiotemporal coherence
Jul 08, 2024Abstract:Super-resolution ultrasound imaging through microbubble (MB) localisation and tracking, also known as ultrasound localisation microscopy, allows non-invasive sub-diffraction resolution imaging of microvasculature in animals and humans. The number of MBs localised from the acquired contrast-enhanced ultrasound (CEUS) images and the localisation precision directly influence the quality of the resulting super-resolution microvasculature images. However, non-negligible noise present in the CEUS images can make localising MBs challenging. To enhance the MB localisation performance, we propose a Multi-Frame Deconvolution (MF-Decon) framework that can exploit the spatiotemporal coherence inherent in the CEUS data, with new spatial and temporal regularisers designed based on total variation (TV) and regularisation by denoising (RED). Based on the MF-Decon framework, we introduce two novel methods: MF-Decon with spatial and temporal TVs (MF-Decon+3DTV) and MF-Decon with spatial RED and temporal TV (MF-Decon+RED+TV). Results from in silico simulations indicate that our methods outperform two widely used methods using deconvolution or normalised cross-correlation across all evaluation metrics, including precision, recall, $F_1$ score, mean and standard localisation errors. In particular, our methods improve MB localisation precision by up to 39% and recall by up to 12%. Super-resolution microvasculature maps generated with our methods on a publicly available in vivo rat brain dataset show less noise, better contrast, higher resolution and more vessel structures.
Ultrafast 3-D Super Resolution Ultrasound using Row-Column Array specific Coherence-based Beamforming and Rolling Acoustic Sub-aperture Processing: In Vitro, In Vivo and Clinical Study
Nov 15, 2023



Abstract:The row-column addressed array is an emerging probe for ultrafast 3-D ultrasound imaging. It achieves this with far fewer independent electronic channels and a wider field of view than traditional 2-D matrix arrays, of the same channel count, making it a good candidate for clinical translation. However, the image quality of row-column arrays is generally poor, particularly when investigating tissue. Ultrasound localisation microscopy allows for the production of super-resolution images even when the initial image resolution is not high. Unfortunately, the row-column probe can suffer from imaging artefacts that can degrade the quality of super-resolution images as `secondary' lobes from bright microbubbles can be mistaken as microbubble events, particularly when operated using plane wave imaging. These false events move through the image in a physiologically realistic way so can be challenging to remove via tracking, leading to the production of 'false vessels'. Here, a new type of rolling window image reconstruction procedure was developed, which integrated a row-column array-specific coherence-based beamforming technique with acoustic sub-aperture processing for the purposes of reducing `secondary' lobe artefacts, noise and increasing the effective frame rate. Using an {\it{in vitro}} cross tube, it was found that the procedure reduced the percentage of `false' locations from $\sim$26\% to $\sim$15\% compared to traditional orthogonal plane wave compounding. Additionally, it was found that the noise could be reduced by $\sim$7 dB and that the effective frame rate could be increased to over 4000 fps. Subsequently, {\it{in vivo}} ultrasound localisation microscopy was used to produce images non-invasively of a rabbit kidney and a human thyroid.
On the Use of Singular Value Decomposition as a Clutter Filter for Ultrasound Flow Imaging
Apr 25, 2023



Abstract:Filtering based on Singular Value Decomposition (SVD) provides substantial separation of clutter, flow and noise in high frame rate ultrasound flow imaging. The use of SVD as a clutter filter has greatly improved techniques such as vector flow imaging, functional ultrasound and super-resolution ultrasound localization microscopy. The removal of clutter and noise relies on the assumption that tissue, flow and noise are each represented by different subsets of singular values, so that their signals are uncorrelated and lay on orthogonal sub-spaces. This assumption fails in the presence of tissue motion, for near-wall or microvascular flow, and can be influenced by an incorrect choice of singular value thresholds. Consequently, separation of flow, clutter and noise is imperfect, which can lead to image artefacts not present in the original data. Temporal and spatial fluctuation in intensity are the commonest artefacts, which vary in appearance and strengths. Ghosting and splitting artefacts are observed in the microvasculature where the flow signal is sparsely distributed. Singular value threshold selection, tissue motion, frame rate, flow signal amplitude and acquisition length affect the prevalence of these artefacts. Understanding what causes artefacts due to SVD clutter and noise removal is necessary for their interpretation.
Acceleration-Based Kalman Tracking for Super-Resolution Ultrasound Imaging in vivo
Apr 03, 2023Abstract:Super-resolution ultrasound can image microvascular structure and flow at sub-wave-diffraction resolution based on localising and tracking microbubbles. Currently, tracking microbubbles accurately under limited imaging frame rates and high microbubble concentrations remains a challenge, especially under the effect of cardiac pulsatility and in highly curved vessels. In this study, an acceleration-incorporated microbubble motion model is introduced into a Kalman tracking framework. The tracking performance was evaluated using simulated microvasculature with different microbubble motion parameters and acquisition frame rates, and in vivo human breast tumour ultrasound datasets. The simulation results show that the acceleration-based method outperformed the non-acceleration-based method at different levels of acceleration and acquisition frame rates and achieved significant improvement in true positive rate (up to 10.03%), false negative rate (up to 28.61%) and correctly pairing fraction (up to 170.14%). The proposed method can also reduce errors in vasculature reconstruction via the acceleration-based nonlinear interpolation, compared with linear interpolation (up to 19 um). The tracking results from temporally downsampled low frame rate in vivo datasets from human breast tumours show that the proposed method has better microbubble tracking performance than the baseline method, if using results from the initial high frame data as reference. Finally, the acceleration estimated from tracking results also provides a spatial speed gradient map that may contain extra valuable diagnostic information.
Transthoracic super-resolution ultrasound localisation microscopy of myocardial vasculature in patients
Mar 28, 2023Abstract:Micro-vascular flow in the myocardium is of significant importance clinically but remains poorly understood. Up to 25% of patients with symptoms of coronary heart diseases have no obstructive coronary arteries and have suspected microvascular diseases. However, such microvasculature is difficult to image in vivo with existing modalities due to the lack of resolution and sensitivity. Here, we demonstrate the feasibility of transthoracic super-resolution ultrasound localisation microscopy (SRUS/ULM) of myocardial microvasculature and hemodynamics in a large animal model and in patients, using a cardiac phased array probe with a customised data acquisition and processing pipeline. A multi-level motion correction strategy was proposed. A tracking framework incorporating multiple features and automatic parameter initialisations was developed to reconstruct microcirculation. In two patients with impaired myocardial function, we have generated SRUS images of myocardial vascular structure and flow with a resolution that is beyond the wave-diffraction limit (half a wavelength), using data acquired within a breath hold. Myocardial SRUS/ULM has potential to improve the understanding of myocardial microcirculation and the management of patients with cardiac microvascular diseases.
BUbble Flow Field: a Simulation Framework for Evaluating Ultrasound Localization Microscopy Algorithms
Nov 01, 2022



Abstract:Ultrasound contrast enhanced imaging has seen widespread uptake in research and clinical diagnostic imaging. This includes applications such as vector flow imaging, functional ultrasound and super-resolution Ultrasound Localization Microscopy (ULM). All of these require testing and validation during development of new algorithms with ground truth data. In this work we present a comprehensive simulation platform BUbble Flow Field (BUFF) that generates contrast enhanced ultrasound images in vascular tree geometries with realistic flow characteristics and validation algorithms for ULM. BUFF allows complex micro-vascular network generation of random and user-defined vascular networks. Blood flow is simulated with a fast Computational Fluid Dynamics (CFD) solver and allows arbitrary input and output positions and custom pressures. The acoustic field simulation is combined with non-linear Microbubble (MB) dynamics and simulates a range of point spread functions based on user-defined MB characteristics. The validation combines both binary and quantitative metrics. BFF's capacity to generate and validate user-defined networks is demonstrated through its implementation in the Ultrasound Localisation and TRacking Algorithms for Super Resolution (ULTRA-SR) Challenge at the International Ultrasonics Symposium (IUS) 2022 of the Institute of Electrical and Electronics Engineers (IEEE). The ability to produce ULM images, and the availability of a ground truth in localisation and tracking enables objective and quantitative evaluation of the large number of localisation and tracking algorithms developed in the field. BUFF can also benefit deep learning based methods by automatically generating datasets for training. BUFF is a fully comprehensive simulation platform for testing and validation of novel ULM techniques and is open source.
3D Super-Resolution Ultrasound with Adaptive Weight-Based Beamforming
Aug 25, 2022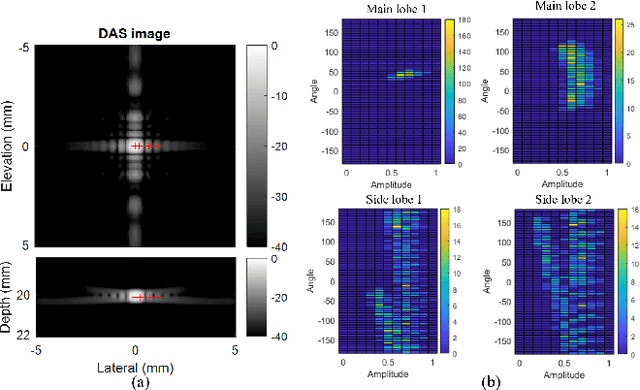
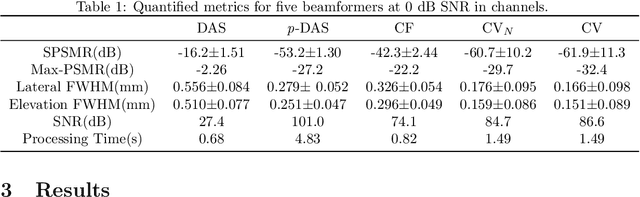
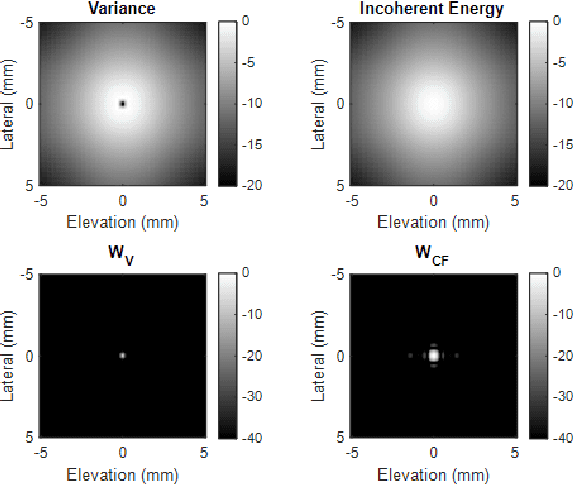
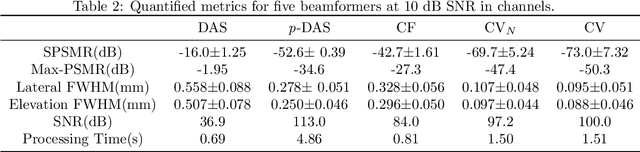
Abstract:Super-resolution ultrasound (SRUS) imaging through localising and tracking sparse microbubbles has been shown to reveal microvascular structure and flow beyond the wave diffraction limit. Most SRUS studies use standard delay and sum (DAS) beamforming, where large main lobe and significant side lobes make separation and localisation of densely distributed bubbles challenging, particularly in 3D due to the typically small aperture of matrix array probes. This study aims to improve 3D SRUS by implementing a low-cost 3D coherence beamformer based on channel signal variance, as well as two other adaptive weight-based coherence beamformers: nonlinear beamforming with p-th root compression and coherence factor. The 3D coherence beamformers, together with DAS, are compared in computer simulation, on a microflow phantom, and in vivo. Simulation results demonstrate that the adaptive weight-based beamformers can significantly narrow the main lobe and suppress the side lobes for modest computational cost. Significantly improved 3D SR images of microflow phantom and a rabbit kidney are obtained through the adaptive weight-based beamformers. The proposed variance-based beamformer performs best in simulations and experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge