Chandni Gupta
Standard Plane Detection in 3D Fetal Ultrasound Using an Iterative Transformation Network
Oct 07, 2018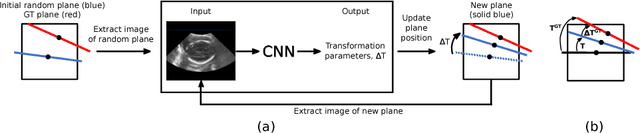
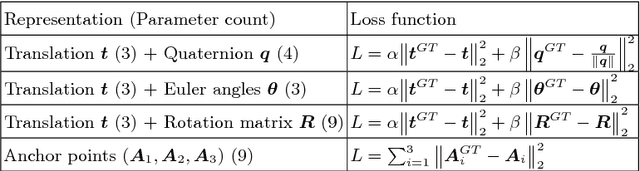
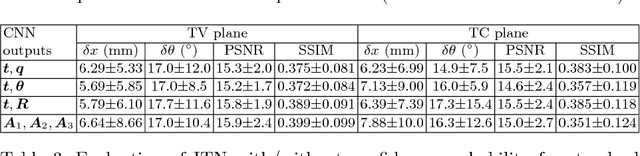
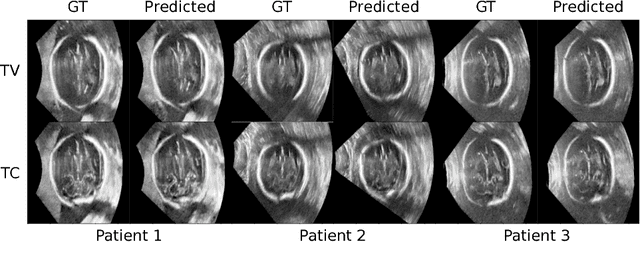
Abstract:Standard scan plane detection in fetal brain ultrasound (US) forms a crucial step in the assessment of fetal development. In clinical settings, this is done by manually manoeuvring a 2D probe to the desired scan plane. With the advent of 3D US, the entire fetal brain volume containing these standard planes can be easily acquired. However, manual standard plane identification in 3D volume is labour-intensive and requires expert knowledge of fetal anatomy. We propose a new Iterative Transformation Network (ITN) for the automatic detection of standard planes in 3D volumes. ITN uses a convolutional neural network to learn the relationship between a 2D plane image and the transformation parameters required to move that plane towards the location/orientation of the standard plane in the 3D volume. During inference, the current plane image is passed iteratively to the network until it converges to the standard plane location. We explore the effect of using different transformation representations as regression outputs of ITN. Under a multi-task learning framework, we introduce additional classification probability outputs to the network to act as confidence measures for the regressed transformation parameters in order to further improve the localisation accuracy. When evaluated on 72 US volumes of fetal brain, our method achieves an error of 3.83mm/12.7 degrees and 3.80mm/12.6 degrees for the transventricular and transcerebellar planes respectively and takes 0.46s per plane. Source code is publicly available at https://github.com/yuanwei1989/plane-detection.
* 8 pages, 2 figures, accepted for MICCAI 2018; Added link to source code
Fast Multiple Landmark Localisation Using a Patch-based Iterative Network
Oct 07, 2018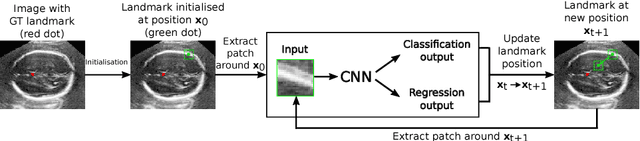
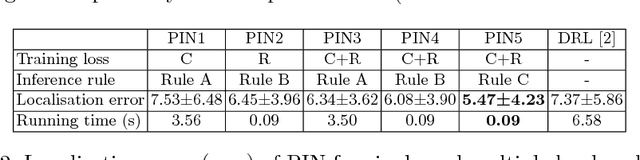
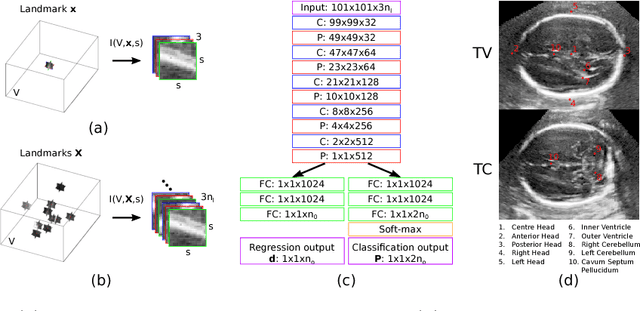
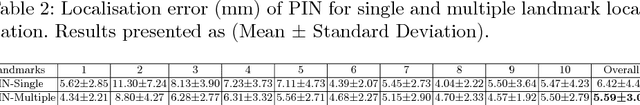
Abstract:We propose a new Patch-based Iterative Network (PIN) for fast and accurate landmark localisation in 3D medical volumes. PIN utilises a Convolutional Neural Network (CNN) to learn the spatial relationship between an image patch and anatomical landmark positions. During inference, patches are repeatedly passed to the CNN until the estimated landmark position converges to the true landmark location. PIN is computationally efficient since the inference stage only selectively samples a small number of patches in an iterative fashion rather than a dense sampling at every location in the volume. Our approach adopts a multi-task learning framework that combines regression and classification to improve localisation accuracy. We extend PIN to localise multiple landmarks by using principal component analysis, which models the global anatomical relationships between landmarks. We have evaluated PIN using 72 3D ultrasound images from fetal screening examinations. PIN achieves quantitatively an average landmark localisation error of 5.59mm and a runtime of 0.44s to predict 10 landmarks per volume. Qualitatively, anatomical 2D standard scan planes derived from the predicted landmark locations are visually similar to the clinical ground truth. Source code is publicly available at https://github.com/yuanwei1989/landmark-detection.
* 8 pages, 4 figures, Accepted for MICCAI 2018
EchoFusion: Tracking and Reconstruction of Objects in 4D Freehand Ultrasound Imaging without External Trackers
Jul 19, 2018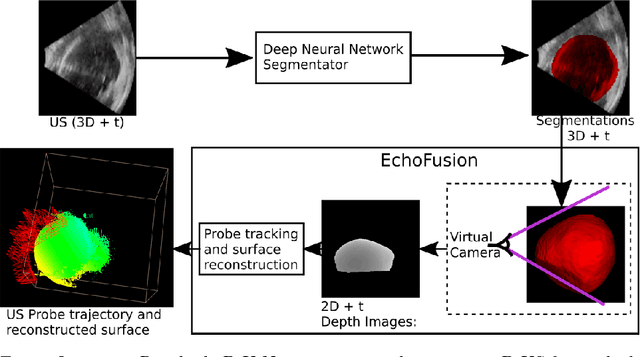

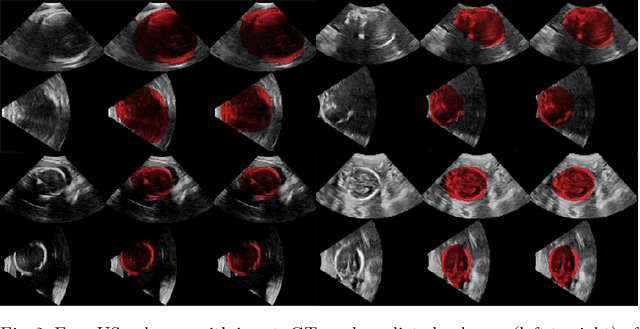
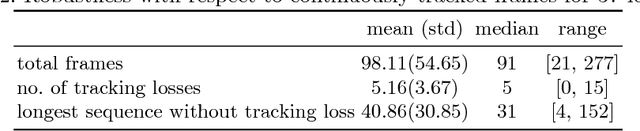
Abstract:Ultrasound (US) is the most widely used fetal imaging technique. However, US images have limited capture range, and suffer from view dependent artefacts such as acoustic shadows. Compounding of overlapping 3D US acquisitions into a high-resolution volume can extend the field of view and remove image artefacts, which is useful for retrospective analysis including population based studies. However, such volume reconstructions require information about relative transformations between probe positions from which the individual volumes were acquired. In prenatal US scans, the fetus can move independently from the mother, making external trackers such as electromagnetic or optical tracking unable to track the motion between probe position and the moving fetus. We provide a novel methodology for image-based tracking and volume reconstruction by combining recent advances in deep learning and simultaneous localisation and mapping (SLAM). Tracking semantics are established through the use of a Residual 3D U-Net and the output is fed to the SLAM algorithm. As a proof of concept, experiments are conducted on US volumes taken from a whole body fetal phantom, and from the heads of real fetuses. For the fetal head segmentation, we also introduce a novel weak annotation approach to minimise the required manual effort for ground truth annotation. We evaluate our method qualitatively, and quantitatively with respect to tissue discrimination accuracy and tracking robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge