Emily Skelton
Placenta Segmentation in Ultrasound Imaging: Addressing Sources of Uncertainty and Limited Field-of-View
Jun 29, 2022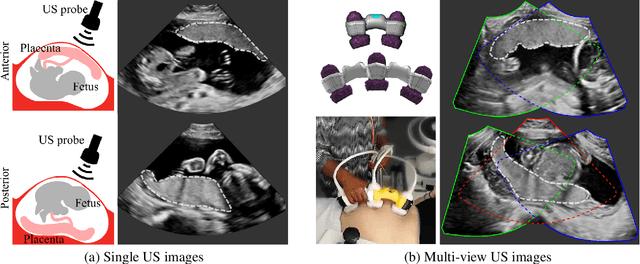
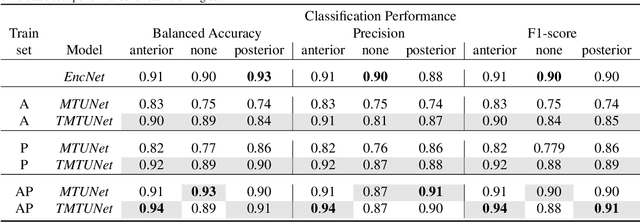
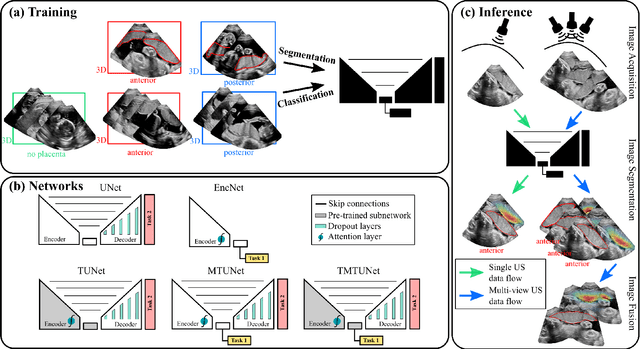
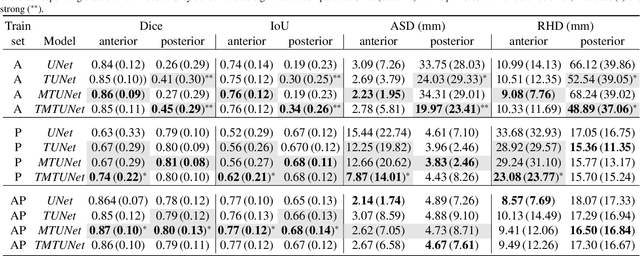
Abstract:Automatic segmentation of the placenta in fetal ultrasound (US) is challenging due to the (i) high diversity of placenta appearance, (ii) the restricted quality in US resulting in highly variable reference annotations, and (iii) the limited field-of-view of US prohibiting whole placenta assessment at late gestation. In this work, we address these three challenges with a multi-task learning approach that combines the classification of placental location (e.g., anterior, posterior) and semantic placenta segmentation in a single convolutional neural network. Through the classification task the model can learn from larger and more diverse datasets while improving the accuracy of the segmentation task in particular in limited training set conditions. With this approach we investigate the variability in annotations from multiple raters and show that our automatic segmentations (Dice of 0.86 for anterior and 0.83 for posterior placentas) achieve human-level performance as compared to intra- and inter-observer variability. Lastly, our approach can deliver whole placenta segmentation using a multi-view US acquisition pipeline consisting of three stages: multi-probe image acquisition, image fusion and image segmentation. This results in high quality segmentation of larger structures such as the placenta in US with reduced image artifacts which are beyond the field-of-view of single probes.
Can non-specialists provide high quality gold standard labels in challenging modalities?
Jul 30, 2021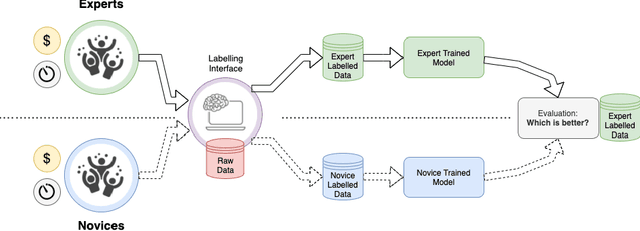
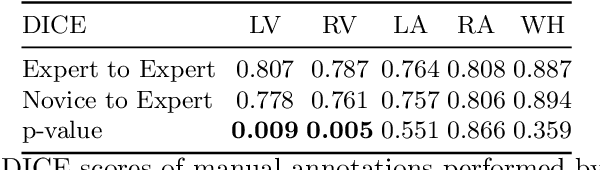
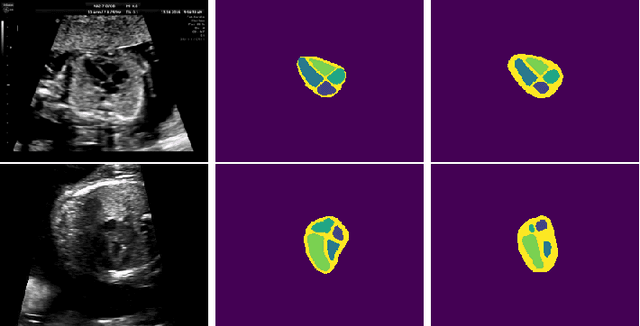
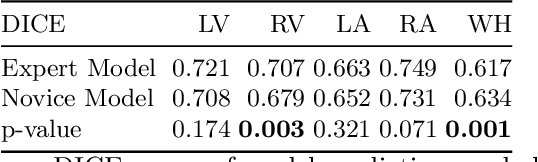
Abstract:Probably yes. -- Supervised Deep Learning dominates performance scores for many computer vision tasks and defines the state-of-the-art. However, medical image analysis lags behind natural image applications. One of the many reasons is the lack of well annotated medical image data available to researchers. One of the first things researchers are told is that we require significant expertise to reliably and accurately interpret and label such data. We see significant inter- and intra-observer variability between expert annotations of medical images. Still, it is a widely held assumption that novice annotators are unable to provide useful annotations for use by clinical Deep Learning models. In this work we challenge this assumption and examine the implications of using a minimally trained novice labelling workforce to acquire annotations for a complex medical image dataset. We study the time and cost implications of using novice annotators, the raw performance of novice annotators compared to gold-standard expert annotators, and the downstream effects on a trained Deep Learning segmentation model's performance for detecting a specific congenital heart disease (hypoplastic left heart syndrome) in fetal ultrasound imaging.
Detecting Hypo-plastic Left Heart Syndrome in Fetal Ultrasound via Disease-specific Atlas Maps
Jul 06, 2021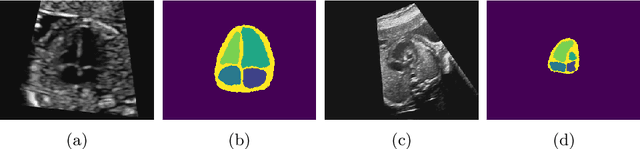
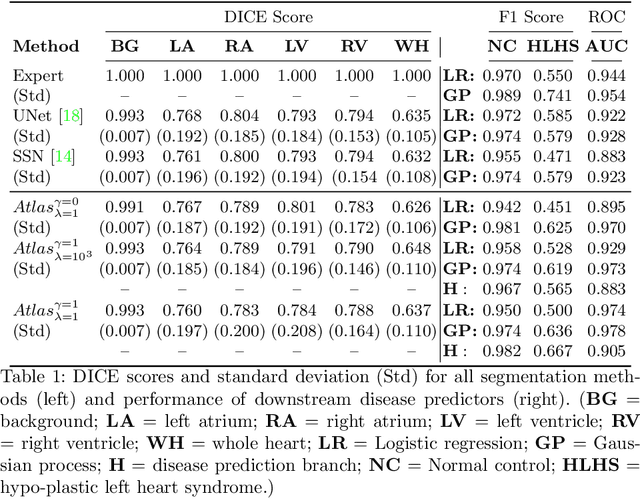
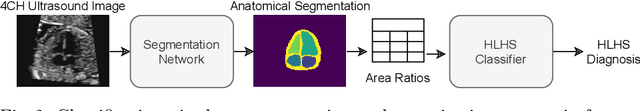
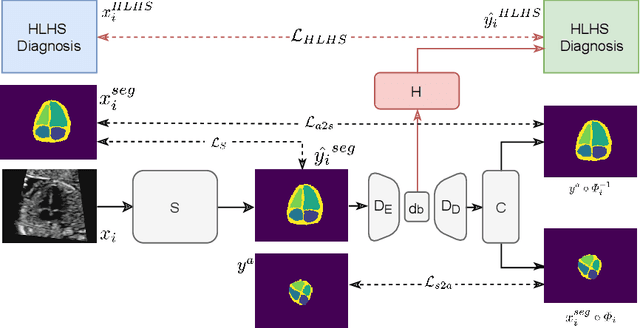
Abstract:Fetal ultrasound screening during pregnancy plays a vital role in the early detection of fetal malformations which have potential long-term health impacts. The level of skill required to diagnose such malformations from live ultrasound during examination is high and resources for screening are often limited. We present an interpretable, atlas-learning segmentation method for automatic diagnosis of Hypo-plastic Left Heart Syndrome (HLHS) from a single `4 Chamber Heart' view image. We propose to extend the recently introduced Image-and-Spatial Transformer Networks (Atlas-ISTN) into a framework that enables sensitising atlas generation to disease. In this framework we can jointly learn image segmentation, registration, atlas construction and disease prediction while providing a maximum level of clinical interpretability compared to direct image classification methods. As a result our segmentation allows diagnoses competitive with expert-derived manual diagnosis and yields an AUC-ROC of 0.978 (1043 cases for training, 260 for validation and 325 for testing).
Screen Tracking for Clinical Translation of Live Ultrasound Image Analysis Methods
Jul 13, 2020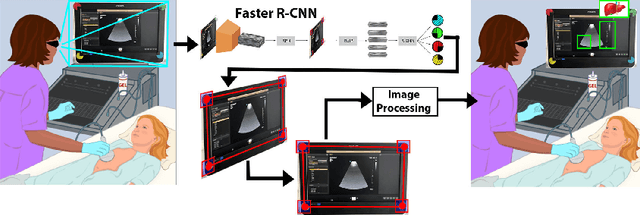
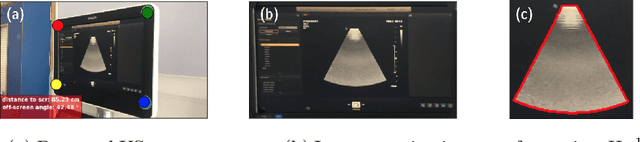
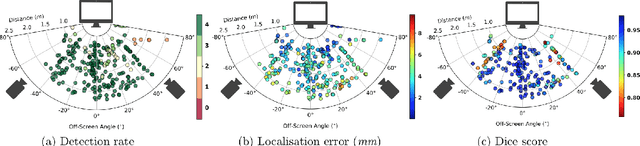
Abstract:Ultrasound (US) imaging is one of the most commonly used non-invasive imaging techniques. However, US image acquisition requires simultaneous guidance of the transducer and interpretation of images, which is a highly challenging task that requires years of training. Despite many recent developments in intra-examination US image analysis, the results are not easy to translate to a clinical setting. We propose a generic framework to extract the US images and superimpose the results of an analysis task, without any need for physical connection or alteration to the US system. The proposed method captures the US image by tracking the screen with a camera fixed at the sonographer's view point and reformats the captured image to the right aspect ratio, in 87.66 +- 3.73ms on average. It is hypothesized that this would enable to input such retrieved image into an image processing pipeline to extract information that can help improve the examination. This information could eventually be projected back to the sonographer's field of view in real time using, for example, an augmented reality (AR) headset.
Robotic-assisted Ultrasound for Fetal Imaging: Evolution from Single-arm to Dual-arm System
Apr 10, 2019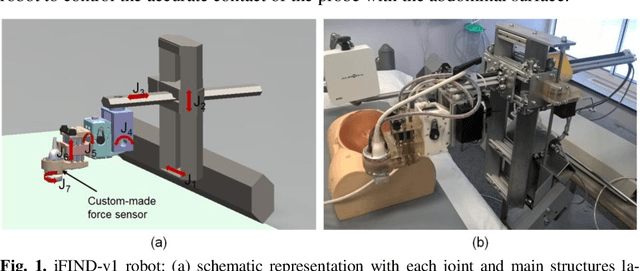
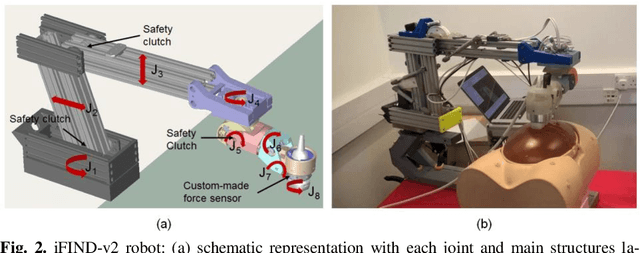
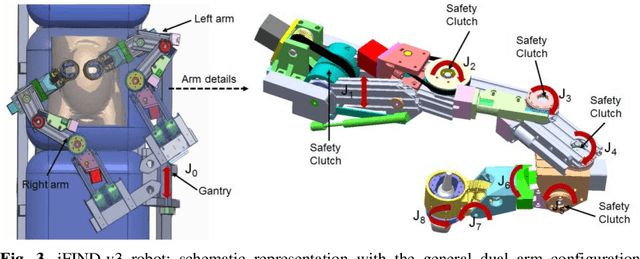
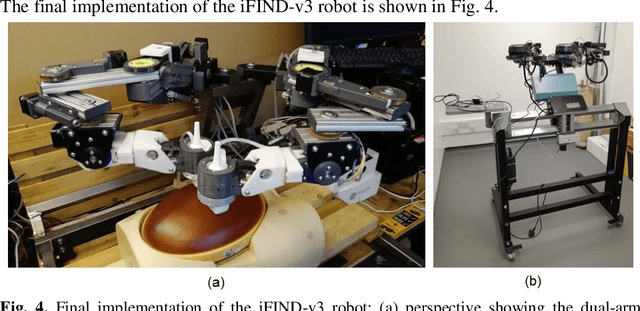
Abstract:The development of robotic-assisted extracorporeal ultrasound systems has a long history and a number of projects have been proposed since the 1990s focusing on different technical aspects. These aim to resolve the deficiencies of on-site manual manipulation of hand-held ultrasound probes. This paper presents the recent ongoing developments of a series of bespoke robotic systems, including both single-arm and dual-arm versions, for a project known as intelligent Fetal Imaging and Diagnosis (iFIND). After a brief review of the development history of the extracorporeal ultrasound robotic system used for fetal and abdominal examinations, the specific aim of the iFIND robots, the design evolution, the implementation details of each version, and the initial clinical feedback of the iFIND robot series are presented. Based on the preliminary testing of these newly-proposed robots on 42 volunteers, the successful and re-liable working of the mechatronic systems were validated. Analysis of a participant questionnaire indicates a comfortable scanning experience for the volunteers and a good acceptance rate to being scanned by the robots.
Weakly Supervised Localisation for Fetal Ultrasound Images
Aug 02, 2018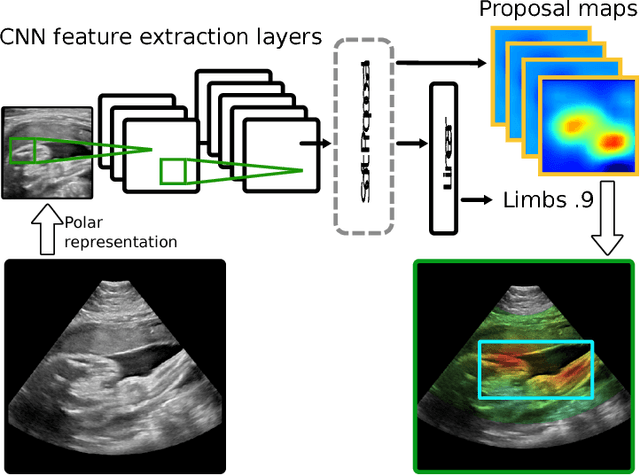
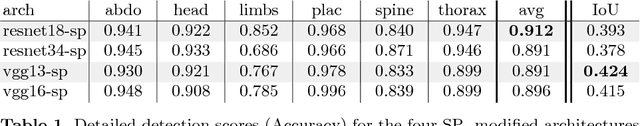
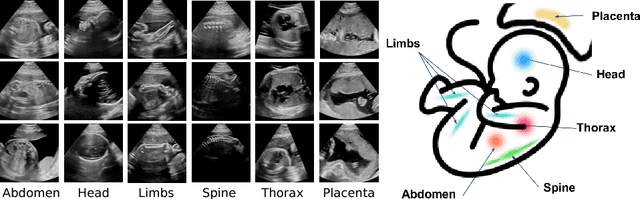
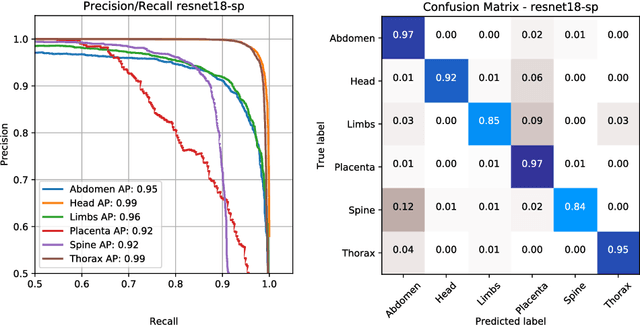
Abstract:This paper addresses the task of detecting and localising fetal anatomical regions in 2D ultrasound images, where only image-level labels are present at training, i.e. without any localisation or segmentation information. We examine the use of convolutional neural network architectures coupled with soft proposal layers. The resulting network simultaneously performs anatomical region detection (classification) and localisation tasks. We generate a proposal map describing the attention of the network for a particular class. The network is trained on 85,500 2D fetal Ultrasound images and their associated labels. Labels correspond to six anatomical regions: head, spine, thorax, abdomen, limbs, and placenta. Detection achieves an average accuracy of 90\% on individual regions, and show that the proposal maps correlate well with relevant anatomical structures. This work presents itself as a powerful and essential step towards subsequent tasks such as fetal position and pose estimation, organ-specific segmentation, or image-guided navigation. Code and additional material is available at https://ntoussaint.github.io/fetalnav
EchoFusion: Tracking and Reconstruction of Objects in 4D Freehand Ultrasound Imaging without External Trackers
Jul 19, 2018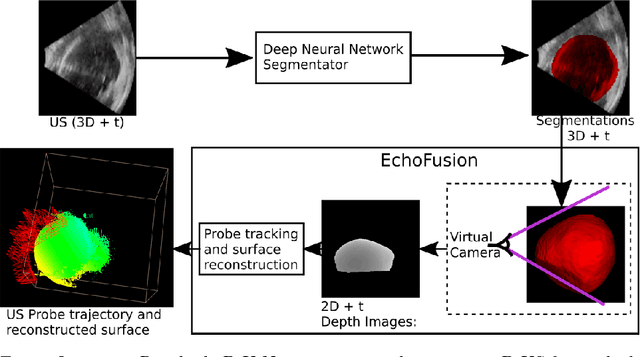

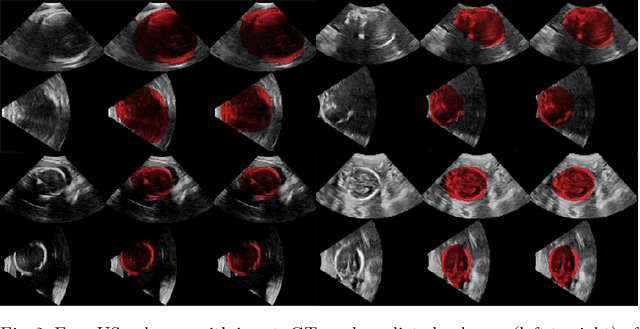
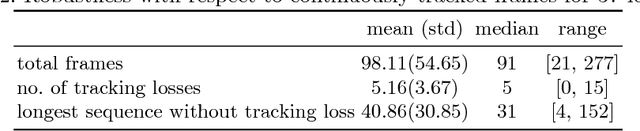
Abstract:Ultrasound (US) is the most widely used fetal imaging technique. However, US images have limited capture range, and suffer from view dependent artefacts such as acoustic shadows. Compounding of overlapping 3D US acquisitions into a high-resolution volume can extend the field of view and remove image artefacts, which is useful for retrospective analysis including population based studies. However, such volume reconstructions require information about relative transformations between probe positions from which the individual volumes were acquired. In prenatal US scans, the fetus can move independently from the mother, making external trackers such as electromagnetic or optical tracking unable to track the motion between probe position and the moving fetus. We provide a novel methodology for image-based tracking and volume reconstruction by combining recent advances in deep learning and simultaneous localisation and mapping (SLAM). Tracking semantics are established through the use of a Residual 3D U-Net and the output is fed to the SLAM algorithm. As a proof of concept, experiments are conducted on US volumes taken from a whole body fetal phantom, and from the heads of real fetuses. For the fetal head segmentation, we also introduce a novel weak annotation approach to minimise the required manual effort for ground truth annotation. We evaluate our method qualitatively, and quantitatively with respect to tissue discrimination accuracy and tracking robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge