Yoshinobu Sato
Division of Information Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, Ikoma city, Japan
Visualization of Organ Movements Using Automatic Region Segmentation of Swallowing CT
Jan 29, 2025



Abstract:This study presents the first report on the development of an artificial intelligence (AI) for automatic region segmentation of four-dimensional computer tomography (4D-CT) images during swallowing. The material consists of 4D-CT images taken during swallowing. Additionally, data for verifying the practicality of the AI were obtained from 4D-CT images during mastication and swallowing. The ground truth data for the region segmentation for the AI were created from five 4D-CT datasets of swallowing. A 3D convolutional model of nnU-Net was used for the AI. The learning and evaluation method for the AI was leave-one-out cross-validation. The number of epochs for training the nnU-Net was 100. The Dice coefficient was used as a metric to assess the AI's region segmentation accuracy. Regions with a median Dice coefficient of 0.7 or higher included the bolus, bones, tongue, and soft palate. Regions with a Dice coefficient below 0.7 included the thyroid cartilage and epiglottis. Factors that reduced the Dice coefficient included metal artifacts caused by dental crowns in the bolus and the speed of movement for the thyroid cartilage and epiglottis. In practical verification of the AI, no significant misrecognition was observed for facial bones, jaw bones, or the tongue. However, regions such as the hyoid bone, thyroid cartilage, and epiglottis were not fully delineated during fast movement. It is expected that future research will improve the accuracy of the AI's region segmentation, though the risk of misrecognition will always exist. Therefore, the development of tools for efficiently correcting the AI's segmentation results is necessary. AI-based visualization is expected to contribute not only to the deepening of motion analysis of organs during swallowing but also to improving the accuracy of swallowing CT by clearly showing the current state of its precision.
3DDX: Bone Surface Reconstruction from a Single Standard-Geometry Radiograph via Dual-Face Depth Estimation
Sep 25, 2024Abstract:Radiography is widely used in orthopedics for its affordability and low radiation exposure. 3D reconstruction from a single radiograph, so-called 2D-3D reconstruction, offers the possibility of various clinical applications, but achieving clinically viable accuracy and computational efficiency is still an unsolved challenge. Unlike other areas in computer vision, X-ray imaging's unique properties, such as ray penetration and fixed geometry, have not been fully exploited. We propose a novel approach that simultaneously learns multiple depth maps (front- and back-surface of multiple bones) derived from the X-ray image to computed tomography registration. The proposed method not only leverages the fixed geometry characteristic of X-ray imaging but also enhances the precision of the reconstruction of the whole surface. Our study involved 600 CT and 2651 X-ray images (4 to 5 posed X-ray images per patient), demonstrating our method's superiority over traditional approaches with a surface reconstruction error reduction from 4.78 mm to 1.96 mm. This significant accuracy improvement and enhanced computational efficiency suggest our approach's potential for clinical application.
Validation of musculoskeletal segmentation model with uncertainty estimation for bone and muscle assessment in hip-to-knee clinical CT images
Sep 04, 2024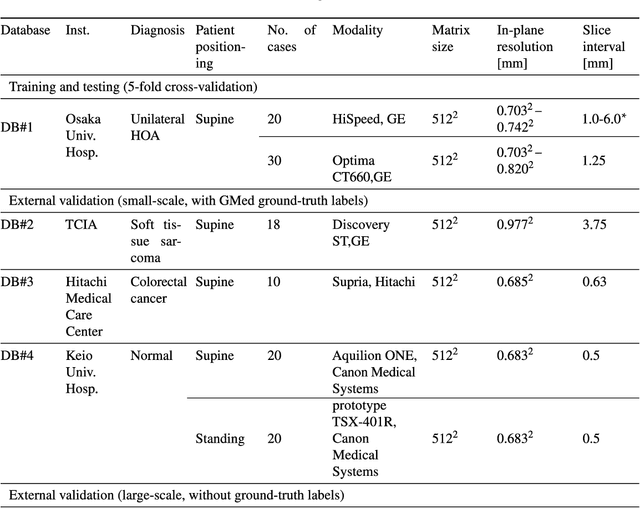

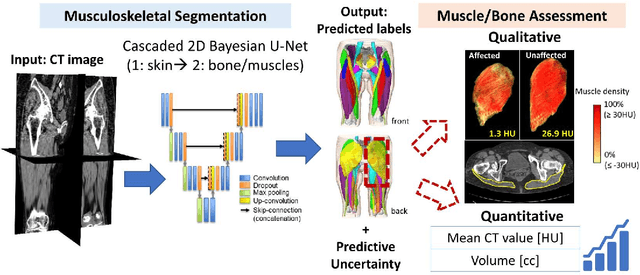
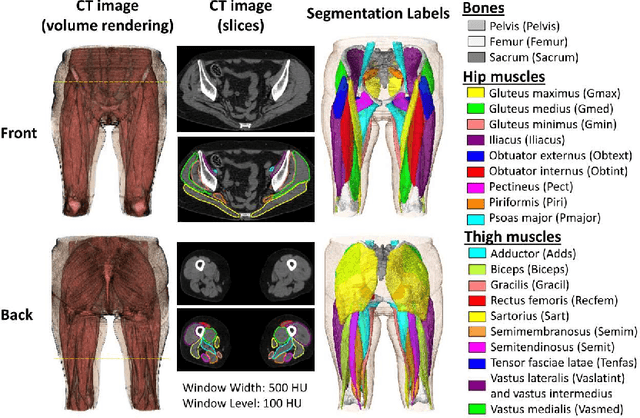
Abstract:Deep learning-based image segmentation has allowed for the fully automated, accurate, and rapid analysis of musculoskeletal (MSK) structures from medical images. However, current approaches were either applied only to 2D cross-sectional images, addressed few structures, or were validated on small datasets, which limit the application in large-scale databases. This study aimed to validate an improved deep learning model for volumetric MSK segmentation of the hip and thigh with uncertainty estimation from clinical computed tomography (CT) images. Databases of CT images from multiple manufacturers/scanners, disease status, and patient positioning were used. The segmentation accuracy, and accuracy in estimating the structures volume and density, i.e., mean HU, were evaluated. An approach for segmentation failure detection based on predictive uncertainty was also investigated. The model has shown an overall improvement with respect to all segmentation accuracy and structure volume/density evaluation metrics. The predictive uncertainty yielded large areas under the receiver operating characteristic (AUROC) curves (AUROCs>=.95) in detecting inaccurate and failed segmentations. The high segmentation and muscle volume/density estimation accuracy, along with the high accuracy in failure detection based on the predictive uncertainty, exhibited the model's reliability for analyzing individual MSK structures in large-scale CT databases.
Enhancing Quantitative Image Synthesis through Pretraining and Resolution Scaling for Bone Mineral Density Estimation from a Plain X-ray Image
Jul 30, 2024Abstract:While most vision tasks are essentially visual in nature (for recognition), some important tasks, especially in the medical field, also require quantitative analysis (for quantification) using quantitative images. Unlike in visual analysis, pixel values in quantitative images correspond to physical metrics measured by specific devices (e.g., a depth image). However, recent work has shown that it is sometimes possible to synthesize accurate quantitative values from visual ones (e.g., depth from visual cues or defocus). This research aims to improve quantitative image synthesis (QIS) by exploring pretraining and image resolution scaling. We propose a benchmark for evaluating pretraining performance using the task of QIS-based bone mineral density (BMD) estimation from plain X-ray images, where the synthesized quantitative image is used to derive BMD. Our results show that appropriate pretraining can improve QIS performance, significantly raising the correlation of BMD estimation from 0.820 to 0.898, while others do not help or even hinder it. Scaling-up the resolution can further boost the correlation up to 0.923, a significant enhancement over conventional methods. Future work will include exploring more pretraining strategies and validating them on other image synthesis tasks.
Automatic hip osteoarthritis grading with uncertainty estimation from computed tomography using digitally-reconstructed radiographs
Dec 30, 2023Abstract:Progression of hip osteoarthritis (hip OA) leads to pain and disability, likely leading to surgical treatment such as hip arthroplasty at the terminal stage. The severity of hip OA is often classified using the Crowe and Kellgren-Lawrence (KL) classifications. However, as the classification is subjective, we aimed to develop an automated approach to classify the disease severity based on the two grades using digitally-reconstructed radiographs (DRRs) from CT images. Automatic grading of the hip OA severity was performed using deep learning-based models. The models were trained to predict the disease grade using two grading schemes, i.e., predicting the Crowe and KL grades separately, and predicting a new ordinal label combining both grades and representing the disease progression of hip OA. The models were trained in classification and regression settings. In addition, the model uncertainty was estimated and validated as a predictor of classification accuracy. The models were trained and validated on a database of 197 hip OA patients, and externally validated on 52 patients. The model accuracy was evaluated using exact class accuracy (ECA), one-neighbor class accuracy (ONCA), and balanced accuracy.The deep learning models produced a comparable accuracy of approximately 0.65 (ECA) and 0.95 (ONCA) in the classification and regression settings. The model uncertainty was significantly larger in cases with large classification errors (P<6e-3). In this study, an automatic approach for grading hip OA severity from CT images was developed. The models have shown comparable performance with high ONCA, which facilitates automated grading in large-scale CT databases and indicates the potential for further disease progression analysis. Classification accuracy was correlated with the model uncertainty, which would allow for the prediction of classification errors.
Hybrid Representation-Enhanced Sampling for Bayesian Active Learning in Musculoskeletal Segmentation of Lower Extremities
Jul 26, 2023Abstract:Purpose: Obtaining manual annotations to train deep learning (DL) models for auto-segmentation is often time-consuming. Uncertainty-based Bayesian active learning (BAL) is a widely-adopted method to reduce annotation efforts. Based on BAL, this study introduces a hybrid representation-enhanced sampling strategy that integrates density and diversity criteria to save manual annotation costs by efficiently selecting the most informative samples. Methods: The experiments are performed on two lower extremity (LE) datasets of MRI and CT images by a BAL framework based on Bayesian U-net. Our method selects uncertain samples with high density and diversity for manual revision, optimizing for maximal similarity to unlabeled instances and minimal similarity to existing training data. We assess the accuracy and efficiency using Dice and a proposed metric called reduced annotation cost (RAC), respectively. We further evaluate the impact of various acquisition rules on BAL performance and design an ablation study for effectiveness estimation. Results: The proposed method showed superiority or non-inferiority to other methods on both datasets across two acquisition rules, and quantitative results reveal the pros and cons of the acquisition rules. Our ablation study in volume-wise acquisition shows that the combination of density and diversity criteria outperforms solely using either of them in musculoskeletal segmentation. Conclusion: Our sampling method is proven efficient in reducing annotation costs in image segmentation tasks. The combination of the proposed method and our BAL framework provides a semi-automatic way for efficient annotation of medical image datasets.
Bone mineral density estimation from a plain X-ray image by learning decomposition into projections of bone-segmented computed tomography
Jul 21, 2023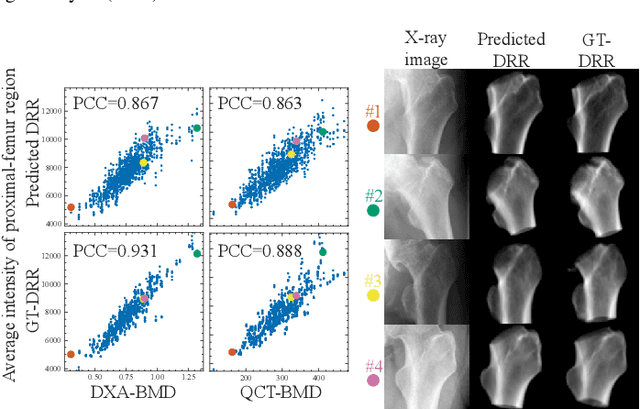
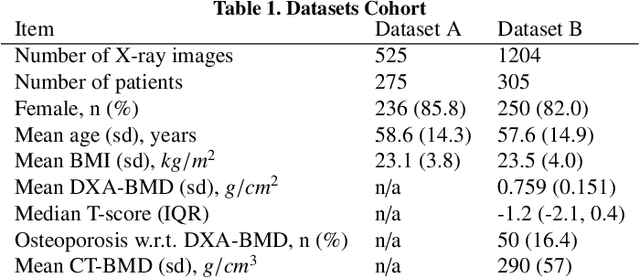
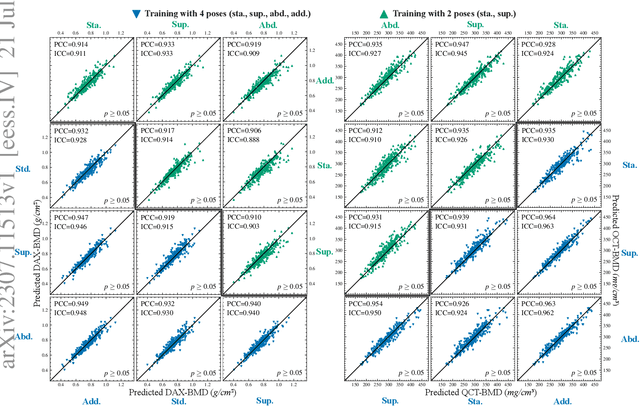
Abstract:Osteoporosis is a prevalent bone disease that causes fractures in fragile bones, leading to a decline in daily living activities. Dual-energy X-ray absorptiometry (DXA) and quantitative computed tomography (QCT) are highly accurate for diagnosing osteoporosis; however, these modalities require special equipment and scan protocols. To frequently monitor bone health, low-cost, low-dose, and ubiquitously available diagnostic methods are highly anticipated. In this study, we aim to perform bone mineral density (BMD) estimation from a plain X-ray image for opportunistic screening, which is potentially useful for early diagnosis. Existing methods have used multi-stage approaches consisting of extraction of the region of interest and simple regression to estimate BMD, which require a large amount of training data. Therefore, we propose an efficient method that learns decomposition into projections of bone-segmented QCT for BMD estimation under limited datasets. The proposed method achieved high accuracy in BMD estimation, where Pearson correlation coefficients of 0.880 and 0.920 were observed for DXA-measured BMD and QCT-measured BMD estimation tasks, respectively, and the root mean square of the coefficient of variation values were 3.27 to 3.79% for four measurements with different poses. Furthermore, we conducted extensive validation experiments, including multi-pose, uncalibrated-CT, and compression experiments toward actual application in routine clinical practice.
MSKdeX: Musculoskeletal (MSK) decomposition from an X-ray image for fine-grained estimation of lean muscle mass and muscle volume
May 31, 2023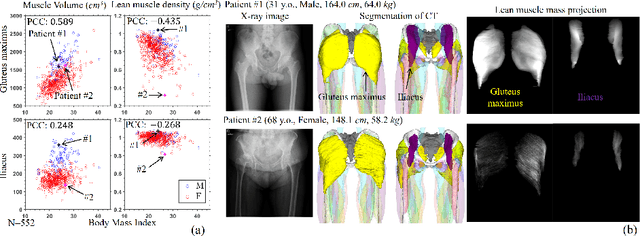
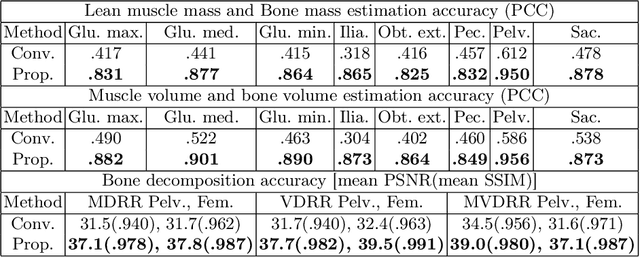
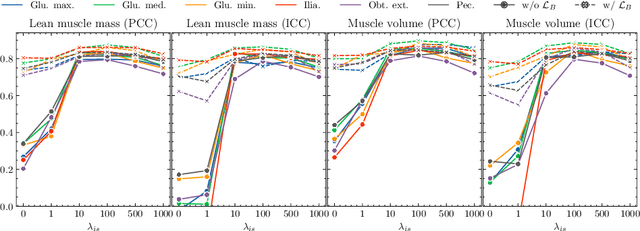
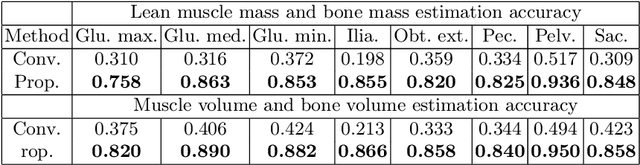
Abstract:Musculoskeletal diseases such as sarcopenia and osteoporosis are major obstacles to health during aging. Although dual-energy X-ray absorptiometry (DXA) and computed tomography (CT) can be used to evaluate musculoskeletal conditions, frequent monitoring is difficult due to the cost and accessibility (as well as high radiation exposure in the case of CT). We propose a method (named MSKdeX) to estimate fine-grained muscle properties from a plain X-ray image, a low-cost, low-radiation, and highly accessible imaging modality, through musculoskeletal decomposition leveraging fine-grained segmentation in CT. We train a multi-channel quantitative image translation model to decompose an X-ray image into projections of CT of individual muscles to infer the lean muscle mass and muscle volume. We propose the object-wise intensity-sum loss, a simple yet surprisingly effective metric invariant to muscle deformation and projection direction, utilizing information in CT and X-ray images collected from the same patient. While our method is basically an unpaired image-to-image translation, we also exploit the nature of the bone's rigidity, which provides the paired data through 2D-3D rigid registration, adding strong pixel-wise supervision in unpaired training. Through the evaluation using a 539-patient dataset, we showed that the proposed method significantly outperformed conventional methods. The average Pearson correlation coefficient between the predicted and CT-derived ground truth metrics was increased from 0.460 to 0.863. We believe our method opened up a new musculoskeletal diagnosis method and has the potential to be extended to broader applications in multi-channel quantitative image translation tasks. Our source code will be released soon.
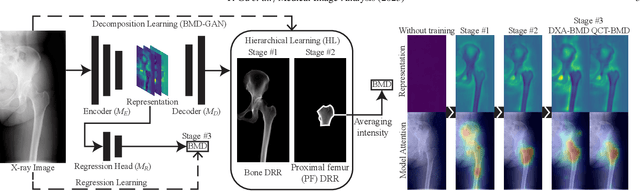
BMD-GAN: Bone mineral density estimation using x-ray image decomposition into projections of bone-segmented quantitative computed tomography using hierarchical learning
Jul 07, 2022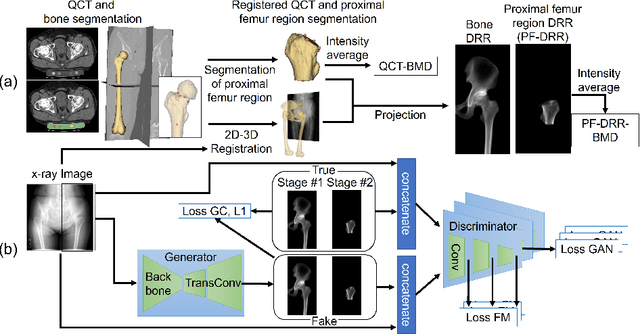
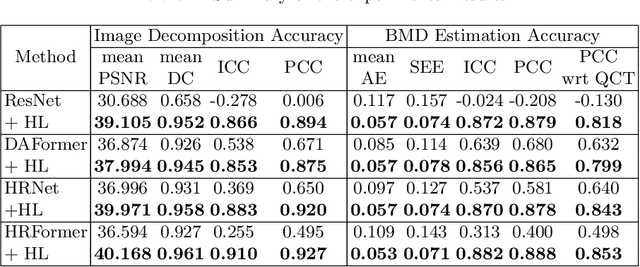
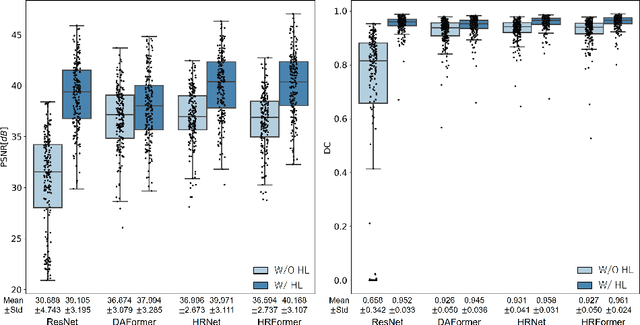
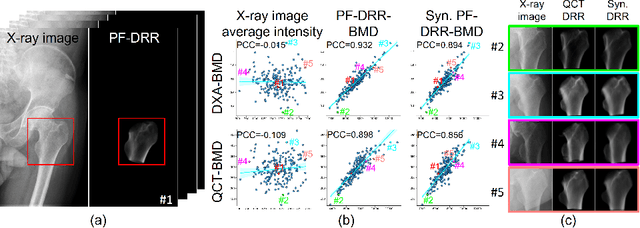
Abstract:We propose a method for estimating the bone mineral density (BMD) from a plain x-ray image. Dual-energy X-ray absorptiometry (DXA) and quantitative computed tomography (QCT) provide high accuracy in diagnosing osteoporosis; however, these modalities require special equipment and scan protocols. Measuring BMD from an x-ray image provides an opportunistic screening, which is potentially useful for early diagnosis. The previous methods that directly learn the relationship between x-ray images and BMD require a large training dataset to achieve high accuracy because of large intensity variations in the x-ray images. Therefore, we propose an approach using the QCT for training a generative adversarial network (GAN) and decomposing an x-ray image into a projection of bone-segmented QCT. The proposed hierarchical learning improved the robustness and accuracy of quantitatively decomposing a small-area target. The evaluation of 200 patients with osteoarthritis using the proposed method, which we named BMD-GAN, demonstrated a Pearson correlation coefficient of 0.888 between the predicted and ground truth DXA-measured BMD. Besides not requiring a large-scale training database, another advantage of our method is its extensibility to other anatomical areas, such as the vertebrae and rib bones.
Automated segmentation of an intensity calibration phantom in clinical CT images using a convolutional neural network
Dec 21, 2020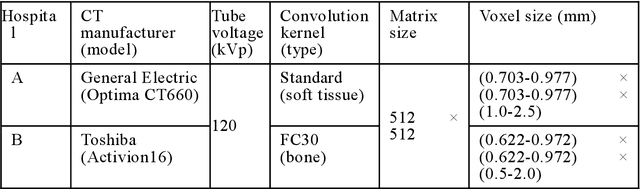
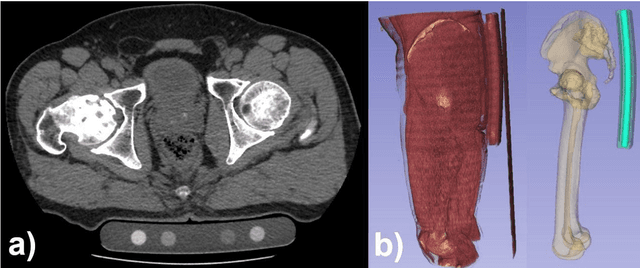
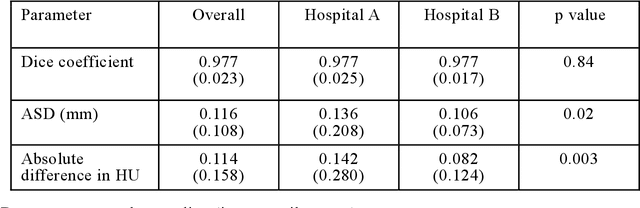
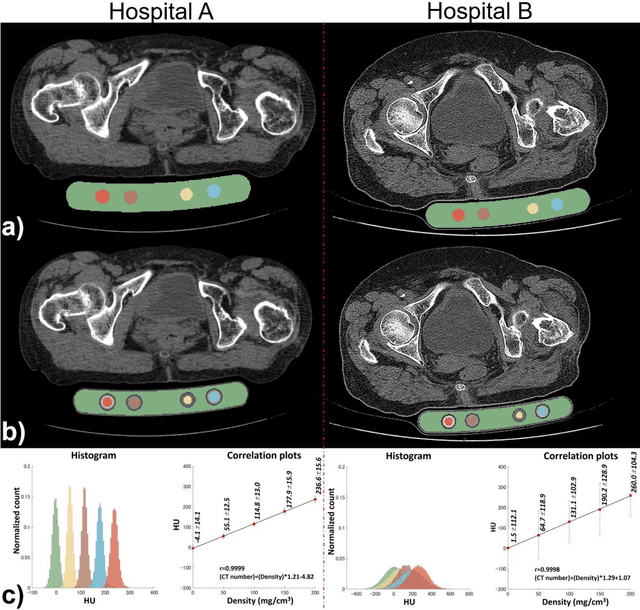
Abstract:Purpose: To apply a convolutional neural network (CNN) to develop a system that segments intensity calibration phantom regions in computed tomography (CT) images, and to test the system in a large cohort to evaluate its robustness. Methods: A total of 1040 cases (520 cases each from two institutions), in which an intensity calibration phantom (B-MAS200, Kyoto Kagaku, Kyoto, Japan) was used, were included herein. A training dataset was created by manually segmenting the regions of the phantom for 40 cases (20 cases each). Segmentation accuracy of the CNN model was assessed with the Dice coefficient and the average symmetric surface distance (ASD) through the 4-fold cross validation. Further, absolute differences of radiodensity values (in Hounsfield units: HU) were compared between manually segmented regions and automatically segmented regions. The system was tested on the remaining 1000 cases. For each institution, linear regression was applied to calculate coefficients for the correlation between radiodensity and the densities of the phantom. Results: After training, the median Dice coefficient was 0.977, and the median ASD was 0.116 mm. When segmented regions were compared between manual segmentation and automated segmentation, the median absolute difference was 0.114 HU. For the test cases, the median correlation coefficient was 0.9998 for one institution and was 0.9999 for the other, with a minimum value of 0.9863. Conclusions: The CNN model successfully segmented the calibration phantom's regions in the CT images with excellent accuracy, and the automated method was found to be at least equivalent to the conventional manual method. Future study should integrate the system by automatically segmenting the region of interest in bones such that the bone mineral density can be fully automatically quantified from CT images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge