Masahiro Jinzaki
EHR-MCP: Real-world Evaluation of Clinical Information Retrieval by Large Language Models via Model Context Protocol
Sep 19, 2025Abstract:Background: Large language models (LLMs) show promise in medicine, but their deployment in hospitals is limited by restricted access to electronic health record (EHR) systems. The Model Context Protocol (MCP) enables integration between LLMs and external tools. Objective: To evaluate whether an LLM connected to an EHR database via MCP can autonomously retrieve clinically relevant information in a real hospital setting. Methods: We developed EHR-MCP, a framework of custom MCP tools integrated with the hospital EHR database, and used GPT-4.1 through a LangGraph ReAct agent to interact with it. Six tasks were tested, derived from use cases of the infection control team (ICT). Eight patients discussed at ICT conferences were retrospectively analyzed. Agreement with physician-generated gold standards was measured. Results: The LLM consistently selected and executed the correct MCP tools. Except for two tasks, all tasks achieved near-perfect accuracy. Performance was lower in the complex task requiring time-dependent calculations. Most errors arose from incorrect arguments or misinterpretation of tool results. Responses from EHR-MCP were reliable, though long and repetitive data risked exceeding the context window. Conclusions: LLMs can retrieve clinical data from an EHR via MCP tools in a real hospital setting, achieving near-perfect performance in simple tasks while highlighting challenges in complex ones. EHR-MCP provides an infrastructure for secure, consistent data access and may serve as a foundation for hospital AI agents. Future work should extend beyond retrieval to reasoning, generation, and clinical impact assessment, paving the way for effective integration of generative AI into clinical practice.
Validation of musculoskeletal segmentation model with uncertainty estimation for bone and muscle assessment in hip-to-knee clinical CT images
Sep 04, 2024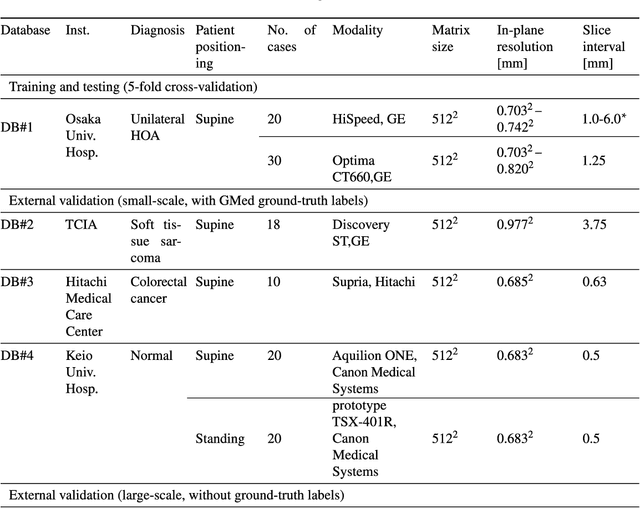

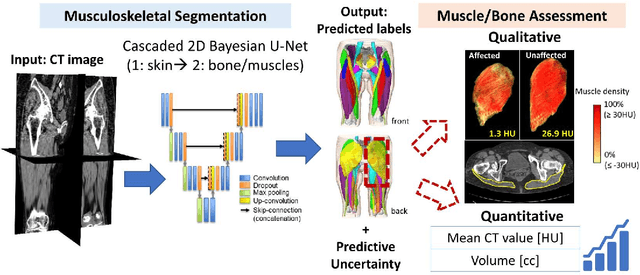
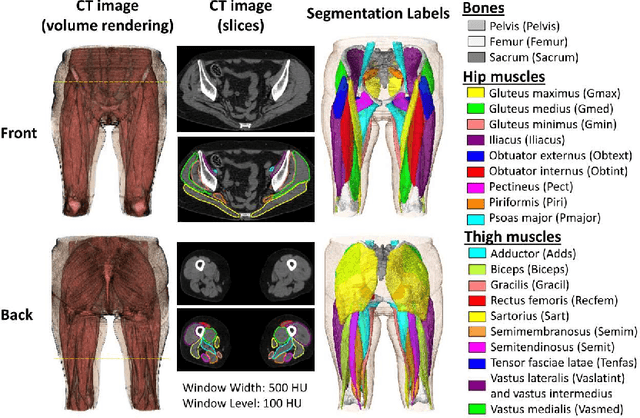
Abstract:Deep learning-based image segmentation has allowed for the fully automated, accurate, and rapid analysis of musculoskeletal (MSK) structures from medical images. However, current approaches were either applied only to 2D cross-sectional images, addressed few structures, or were validated on small datasets, which limit the application in large-scale databases. This study aimed to validate an improved deep learning model for volumetric MSK segmentation of the hip and thigh with uncertainty estimation from clinical computed tomography (CT) images. Databases of CT images from multiple manufacturers/scanners, disease status, and patient positioning were used. The segmentation accuracy, and accuracy in estimating the structures volume and density, i.e., mean HU, were evaluated. An approach for segmentation failure detection based on predictive uncertainty was also investigated. The model has shown an overall improvement with respect to all segmentation accuracy and structure volume/density evaluation metrics. The predictive uncertainty yielded large areas under the receiver operating characteristic (AUROC) curves (AUROCs>=.95) in detecting inaccurate and failed segmentations. The high segmentation and muscle volume/density estimation accuracy, along with the high accuracy in failure detection based on the predictive uncertainty, exhibited the model's reliability for analyzing individual MSK structures in large-scale CT databases.
Semantic Segmentation of Thigh Muscle using 2.5D Deep Learning Network Trained with Limited Datasets
Nov 21, 2019



Abstract:Purpose: We propose a 2.5D deep learning neural network (DLNN) to automatically classify thigh muscle into 11 classes and evaluate its classification accuracy over 2D and 3D DLNN when trained with limited datasets. Enables operator invariant quantitative assessment of the thigh muscle volume change with respect to the disease progression. Materials and methods: Retrospective datasets consist of 48 thigh volume (TV) cropped from CT DICOM images. Cropped volumes were aligned with femur axis and resample in 2 mm voxel-spacing. Proposed 2.5D DLNN consists of three 2D U-Net trained with axial, coronal and sagittal muscle slices respectively. A voting algorithm was used to combine the output of U-Nets to create final segmentation. 2.5D U-Net was trained on PC with 38 TV and the remaining 10 TV were used to evaluate segmentation accuracy of 10 classes within Thigh. The result segmentation of both left and right thigh were de-cropped to original CT volume space. Finally, segmentation accuracies were compared between proposed DLNN and 2D/3D U-Net. Results: Average segmentation DSC score accuracy of all classes with 2.5D U-Net as 91.18% and Average Surface distance (ASD) accuracy as 0.84 mm. We found, mean DSC score for 2D U-Net was 3.3% lower than the that of 2.5D U-Net and mean DSC score of 3D U-Net was 5.7% lower than that of 2.5D U-Net when trained with same datasets. Conclusion: We achieved a faster computationally efficient and automatic segmentation of thigh muscle into 11 classes with reasonable accuracy. Enables quantitative evaluation of muscle atrophy with disease progression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge