Yuta Hiasa
MSKdeX: Musculoskeletal (MSK) decomposition from an X-ray image for fine-grained estimation of lean muscle mass and muscle volume
May 31, 2023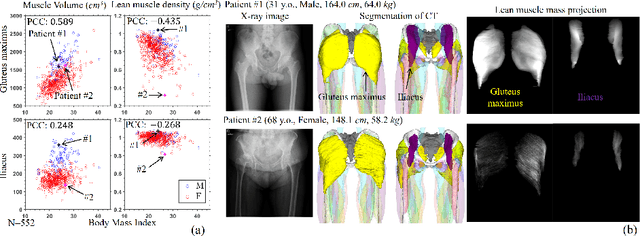
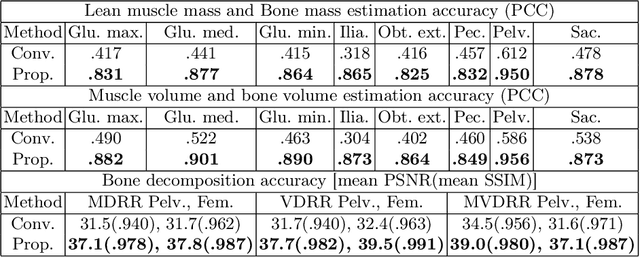
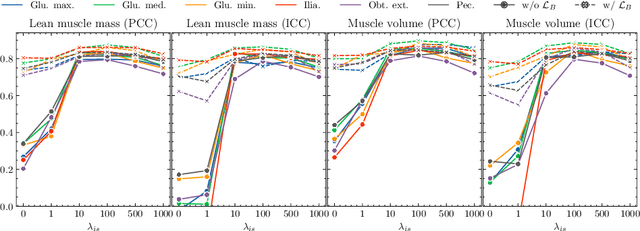
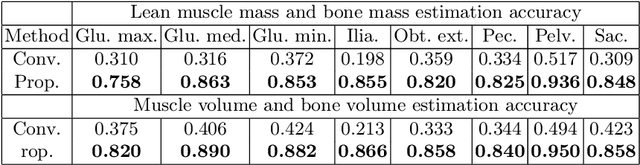
Abstract:Musculoskeletal diseases such as sarcopenia and osteoporosis are major obstacles to health during aging. Although dual-energy X-ray absorptiometry (DXA) and computed tomography (CT) can be used to evaluate musculoskeletal conditions, frequent monitoring is difficult due to the cost and accessibility (as well as high radiation exposure in the case of CT). We propose a method (named MSKdeX) to estimate fine-grained muscle properties from a plain X-ray image, a low-cost, low-radiation, and highly accessible imaging modality, through musculoskeletal decomposition leveraging fine-grained segmentation in CT. We train a multi-channel quantitative image translation model to decompose an X-ray image into projections of CT of individual muscles to infer the lean muscle mass and muscle volume. We propose the object-wise intensity-sum loss, a simple yet surprisingly effective metric invariant to muscle deformation and projection direction, utilizing information in CT and X-ray images collected from the same patient. While our method is basically an unpaired image-to-image translation, we also exploit the nature of the bone's rigidity, which provides the paired data through 2D-3D rigid registration, adding strong pixel-wise supervision in unpaired training. Through the evaluation using a 539-patient dataset, we showed that the proposed method significantly outperformed conventional methods. The average Pearson correlation coefficient between the predicted and CT-derived ground truth metrics was increased from 0.460 to 0.863. We believe our method opened up a new musculoskeletal diagnosis method and has the potential to be extended to broader applications in multi-channel quantitative image translation tasks. Our source code will be released soon.
Region-based Convolution Neural Network Approach for Accurate Segmentation of Pelvic Radiograph
Oct 29, 2019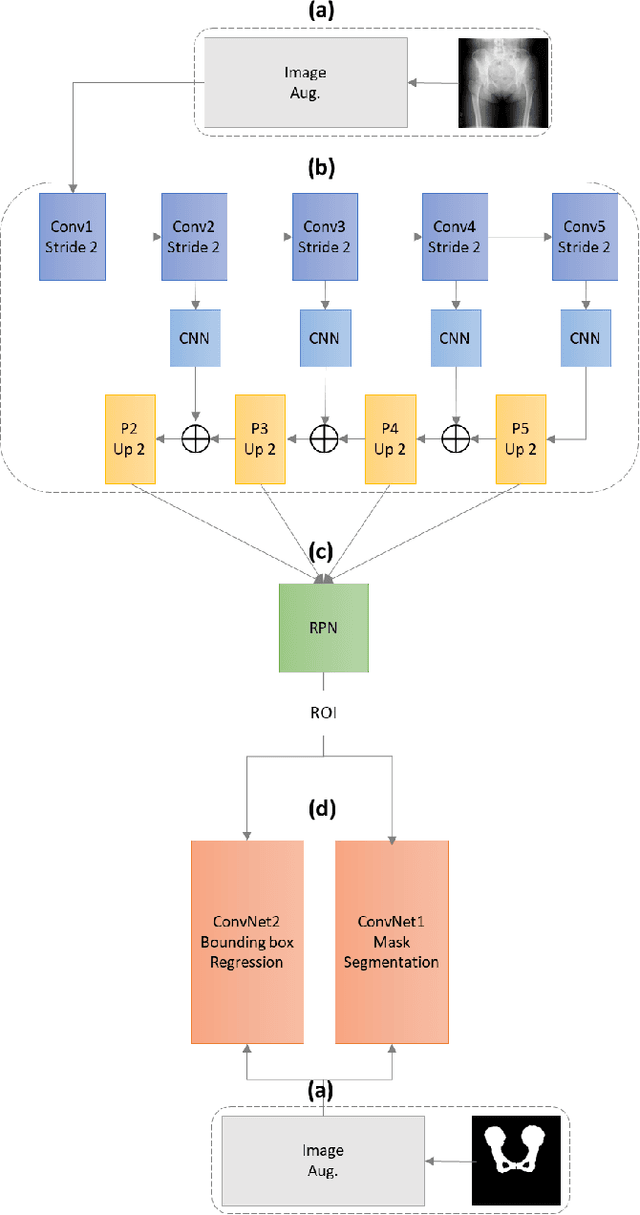
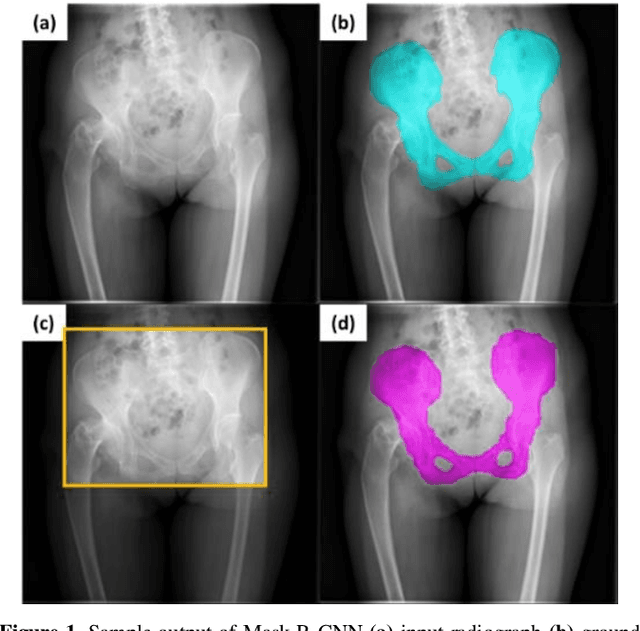
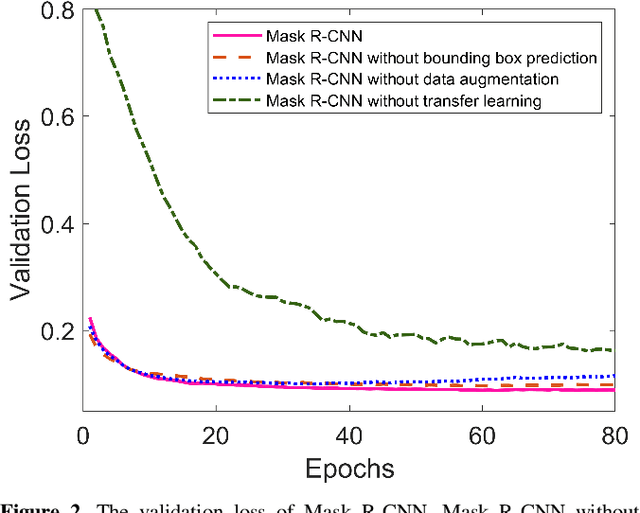
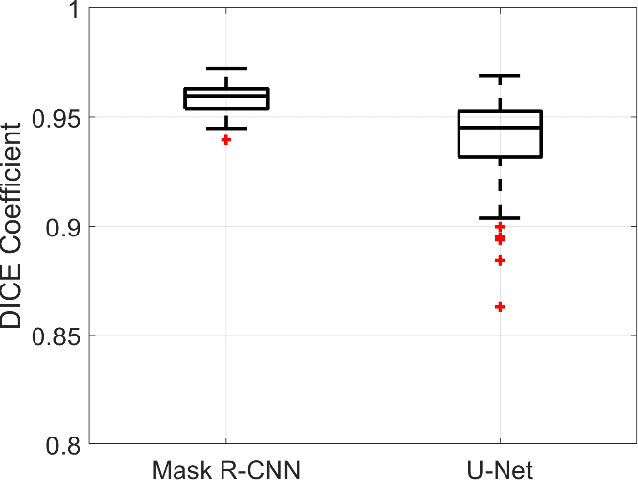
Abstract:With the increasing usage of radiograph images as a most common medical imaging system for diagnosis, treatment planning, and clinical studies, it is increasingly becoming a vital factor to use machine learning-based systems to provide reliable information for surgical pre-planning. Segmentation of pelvic bone in radiograph images is a critical preprocessing step for some applications such as automatic pose estimation and disease detection. However, the encoder-decoder style network known as U-Net has demonstrated limited results due to the challenging complexity of the pelvic shapes, especially in severe patients. In this paper, we propose a novel multi-task segmentation method based on Mask R-CNN architecture. For training, the network weights were initialized by large non-medical dataset and fine-tuned with radiograph images. Furthermore, in the training process, augmented data was generated to improve network performance. Our experiments show that Mask R-CNN utilizing multi-task learning, transfer learning, and data augmentation techniques achieve 0.96 DICE coefficient, which significantly outperforms the U-Net. Notably, for a fair comparison, the same transfer learning and data augmentation techniques have been used for U-net training.
Estimation of Pelvic Sagittal Inclination from Anteroposterior Radiograph Using Convolutional Neural Networks: Proof-of-Concept Study
Oct 26, 2019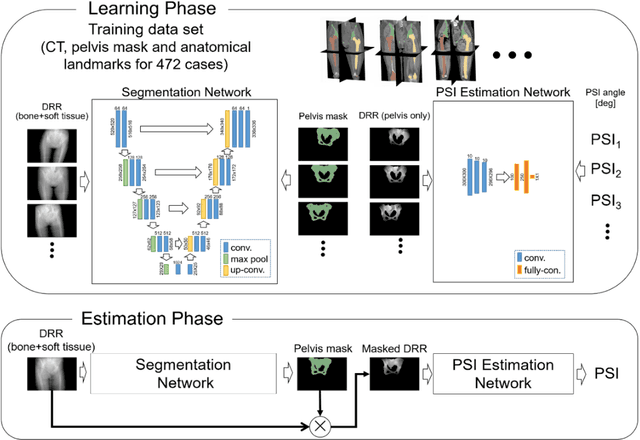
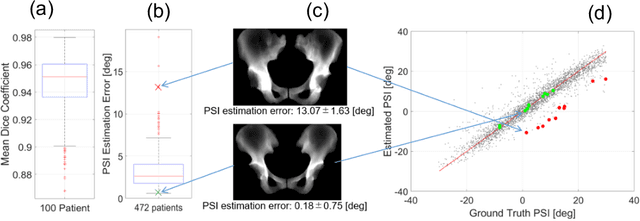
Abstract:Alignment of the bones in standing position provides useful information in surgical planning. In total hip arthroplasty (THA), pelvic sagittal inclination (PSI) angle in the standing position is an important factor in planning of cup alignment and has been estimated mainly from radiographs. Previous methods for PSI estimation used a patient-specific CT to create digitally reconstructed radiographs (DRRs) and compare them with the radiograph to estimate relative position between the pelvis and the x-ray detector. In this study, we developed a method that estimates PSI angle from a single anteroposterior radiograph using two convolutional neural networks (CNNs) without requiring the patient-specific CT, which reduces radiation exposure of the patient and opens up the possibility of application in a larger number of hospitals where CT is not acquired in a routine protocol.
Automated Muscle Segmentation from Clinical CT using Bayesian U-Net for Personalization of a Musculoskeletal Model
Jul 21, 2019



Abstract:We propose a method for automatic segmentation of individual muscles from a clinical CT. The method uses Bayesian convolutional neural networks with the U-Net architecture, using Monte Carlo dropout that infers an uncertainty metric in addition to the segmentation label. We evaluated the performance of the proposed method using two data sets: 20 fully annotated CTs of the hip and thigh regions and 18 partially annotated CTs that are publicly available from The Cancer Imaging Archive (TCIA) database. The experiments showed a Dice coefficient (DC) of 0.891 +/- 0.016 (mean +/- std) and an average symmetric surface distance (ASD) of 0.994 +/- 0.230 mm over 19 muscles in the set of 20 CTs. These results were statistically significant improvements compared to the state-of-the-art hierarchical multi-atlas method which resulted in 0.845 +/- 0.031 DC and 1.556 +/- 0.444 mm ASD. We evaluated validity of the uncertainty metric in the multi-class organ segmentation problem and demonstrated a correlation between the pixels with high uncertainty and the segmentation failure. One application of the uncertainty metric in active-learning is demonstrated, and the proposed query pixel selection method considerably reduced the manual annotation cost for expanding the training data set. The proposed method allows an accurate patient-specific analysis of individual muscle shapes in a clinical routine. This would open up various applications including personalization of biomechanical simulation and quantitative evaluation of muscle atrophy.
Automated Segmentation of Hip and Thigh Muscles in Metal Artifact-Contaminated CT using Convolutional Neural Network-Enhanced Normalized Metal Artifact Reduction
Jun 27, 2019



Abstract:In total hip arthroplasty, analysis of postoperative medical images is important to evaluate surgical outcome. Since Computed Tomography (CT) is most prevalent modality in orthopedic surgery, we aimed at the analysis of CT image. In this work, we focus on the metal artifact in postoperative CT caused by the metallic implant, which reduces the accuracy of segmentation especially in the vicinity of the implant. Our goal was to develop an automated segmentation method of the bones and muscles in the postoperative CT images. We propose a method that combines Normalized Metal Artifact Reduction (NMAR), which is one of the state-of-the-art metal artifact reduction methods, and a Convolutional Neural Network-based segmentation using two U-net architectures. The first U-net refines the result of NMAR and the muscle segmentation is performed by the second U-net. We conducted experiments using simulated images of 20 patients and real images of three patients to evaluate the segmentation accuracy of 19 muscles. In simulation study, the proposed method showed statistically significant improvement (p<0.05) in the average symmetric surface distance (ASD) metric for 14 muscles out of 19 muscles and the average ASD of all muscles from 1.17 +/- 0.543 mm (mean +/- std over all patients) to 1.10 +/- 0.509 mm over our previous method. The real image study using the manual trace of gluteus maximus and medius muscles showed ASD of 1.32 +/- 0.25 mm. Our future work includes training of a network in an end-to-end manner for both the metal artifact reduction and muscle segmentation.
Cross-modality image synthesis from unpaired data using CycleGAN: Effects of gradient consistency loss and training data size
Jul 31, 2018



Abstract:CT is commonly used in orthopedic procedures. MRI is used along with CT to identify muscle structures and diagnose osteonecrosis due to its superior soft tissue contrast. However, MRI has poor contrast for bone structures. Clearly, it would be helpful if a corresponding CT were available, as bone boundaries are more clearly seen and CT has standardized (i.e., Hounsfield) units. Therefore, we aim at MR-to-CT synthesis. The CycleGAN was successfully applied to unpaired CT and MR images of the head, these images do not have as much variation of intensity pairs as do images in the pelvic region due to the presence of joints and muscles. In this paper, we extended the CycleGAN approach by adding the gradient consistency loss to improve the accuracy at the boundaries. We conducted two experiments. To evaluate image synthesis, we investigated dependency of image synthesis accuracy on 1) the number of training data and 2) the gradient consistency loss. To demonstrate the applicability of our method, we also investigated a segmentation accuracy on synthesized images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge