Data Alchemy: Mitigating Cross-Site Model Variability Through Test Time Data Calibration
Paper and Code
Jul 18, 2024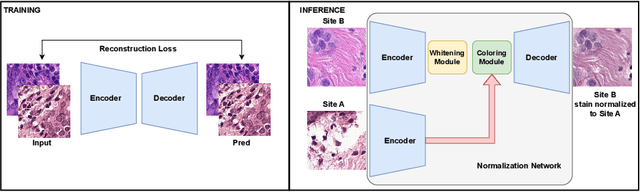
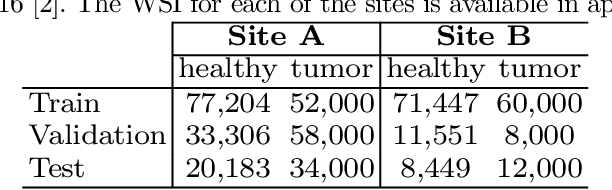
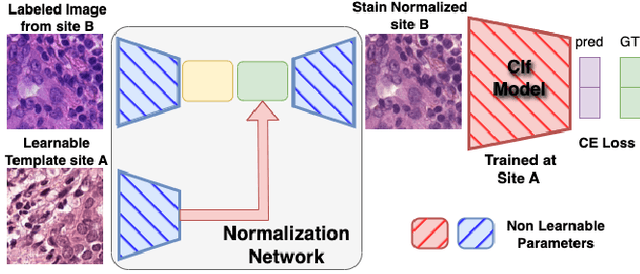
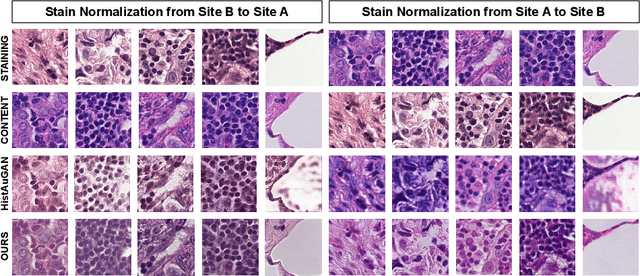
Deploying deep learning-based imaging tools across various clinical sites poses significant challenges due to inherent domain shifts and regulatory hurdles associated with site-specific fine-tuning. For histopathology, stain normalization techniques can mitigate discrepancies, but they often fall short of eliminating inter-site variations. Therefore, we present Data Alchemy, an explainable stain normalization method combined with test time data calibration via a template learning framework to overcome barriers in cross-site analysis. Data Alchemy handles shifts inherent to multi-site data and minimizes them without needing to change the weights of the normalization or classifier networks. Our approach extends to unseen sites in various clinical settings where data domain discrepancies are unknown. Extensive experiments highlight the efficacy of our framework in tumor classification in hematoxylin and eosin-stained patches. Our explainable normalization method boosts classification tasks' area under the precision-recall curve(AUPR) by 0.165, 0.545 to 0.710. Additionally, Data Alchemy further reduces the multisite classification domain gap, by improving the 0.710 AUPR an additional 0.142, elevating classification performance further to 0.852, from 0.545. Our Data Alchemy framework can popularize precision medicine with minimal operational overhead by allowing for the seamless integration of pre-trained deep learning-based clinical tools across multiple sites.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge