Daniel Capellán-Martín
Improving Pre-trained Segmentation Models using Post-Processing
Dec 16, 2025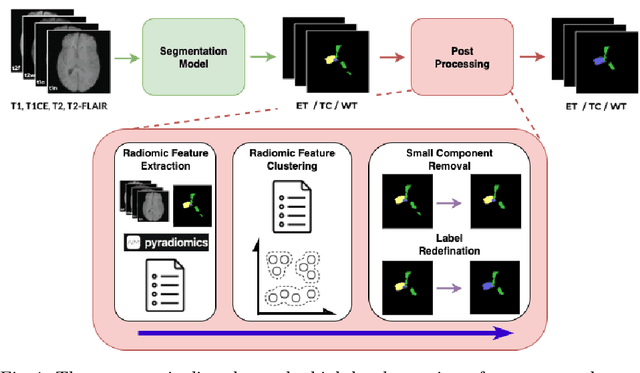

Abstract:Gliomas are the most common malignant brain tumors in adults and are among the most lethal. Despite aggressive treatment, the median survival rate is less than 15 months. Accurate multiparametric MRI (mpMRI) tumor segmentation is critical for surgical planning, radiotherapy, and disease monitoring. While deep learning models have improved the accuracy of automated segmentation, large-scale pre-trained models generalize poorly and often underperform, producing systematic errors such as false positives, label swaps, and slice discontinuities in slices. These limitations are further compounded by unequal access to GPU resources and the growing environmental cost of large-scale model training. In this work, we propose adaptive post-processing techniques to refine the quality of glioma segmentations produced by large-scale pretrained models developed for various types of tumors. We demonstrated the techniques in multiple BraTS 2025 segmentation challenge tasks, with the ranking metric improving by 14.9 % for the sub-Saharan Africa challenge and 0.9% for the adult glioma challenge. This approach promotes a shift in brain tumor segmentation research from increasingly complex model architectures to efficient, clinically aligned post-processing strategies that are precise, computationally fair, and sustainable.
Adaptable Segmentation Pipeline for Diverse Brain Tumors with Radiomic-guided Subtyping and Lesion-Wise Model Ensemble
Dec 16, 2025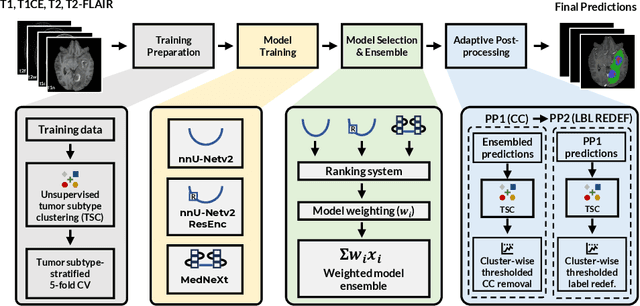
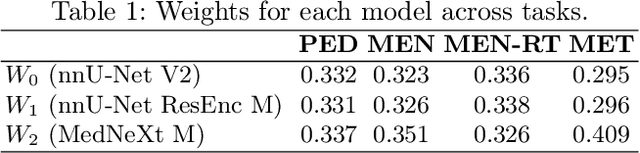
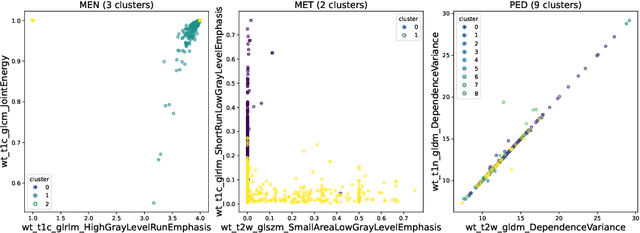
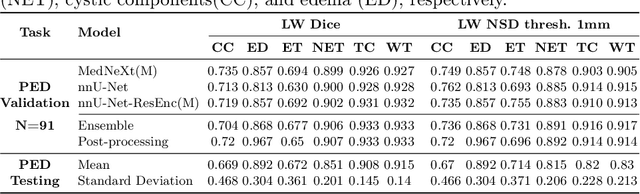
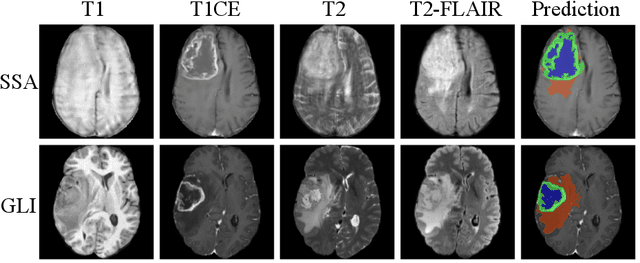
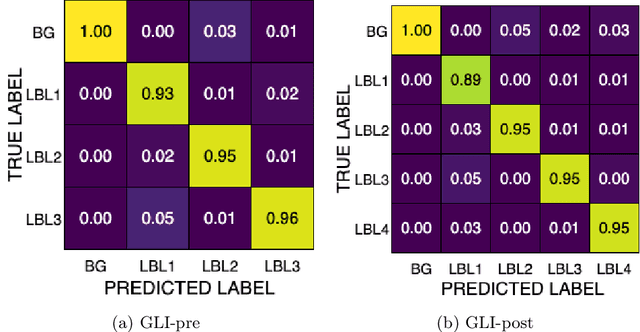
Abstract:Robust and generalizable segmentation of brain tumors on multi-parametric magnetic resonance imaging (MRI) remains difficult because tumor types differ widely. The BraTS 2025 Lighthouse Challenge benchmarks segmentation methods on diverse high-quality datasets of adult and pediatric tumors: multi-consortium international pediatric brain tumor segmentation (PED), preoperative meningioma tumor segmentation (MEN), meningioma radiotherapy segmentation (MEN-RT), and segmentation of pre- and post-treatment brain metastases (MET). We present a flexible, modular, and adaptable pipeline that improves segmentation performance by selecting and combining state-of-the-art models and applying tumor- and lesion-specific processing before and after training. Radiomic features extracted from MRI help detect tumor subtype, ensuring a more balanced training. Custom lesion-level performance metrics determine the influence of each model in the ensemble and optimize post-processing that further refines the predictions, enabling the workflow to tailor every step to each case. On the BraTS testing sets, our pipeline achieved performance comparable to top-ranked algorithms across multiple challenges. These findings confirm that custom lesion-aware processing and model selection yield robust segmentations yet without locking the method to a specific network architecture. Our method has the potential for quantitative tumor measurement in clinical practice, supporting diagnosis and prognosis.
Adult Glioma Segmentation in Sub-Saharan Africa using Transfer Learning on Stratified Finetuning Data
Dec 05, 2024



Abstract:Gliomas, a kind of brain tumor characterized by high mortality, present substantial diagnostic challenges in low- and middle-income countries, particularly in Sub-Saharan Africa. This paper introduces a novel approach to glioma segmentation using transfer learning to address challenges in resource-limited regions with minimal and low-quality MRI data. We leverage pre-trained deep learning models, nnU-Net and MedNeXt, and apply a stratified fine-tuning strategy using the BraTS2023-Adult-Glioma and BraTS-Africa datasets. Our method exploits radiomic analysis to create stratified training folds, model training on a large brain tumor dataset, and transfer learning to the Sub-Saharan context. A weighted model ensembling strategy and adaptive post-processing are employed to enhance segmentation accuracy. The evaluation of our proposed method on unseen validation cases on the BraTS-Africa 2024 task resulted in lesion-wise mean Dice scores of 0.870, 0.865, and 0.926, for enhancing tumor, tumor core, and whole tumor regions and was ranked first for the challenge. Our approach highlights the ability of integrated machine-learning techniques to bridge the gap between the medical imaging capabilities of resource-limited countries and established developed regions. By tailoring our methods to a target population's specific needs and constraints, we aim to enhance diagnostic capabilities in isolated environments. Our findings underscore the importance of approaches like local data integration and stratification refinement to address healthcare disparities, ensure practical applicability, and enhance impact.
Magnetic Resonance Imaging Feature-Based Subtyping and Model Ensemble for Enhanced Brain Tumor Segmentation
Dec 05, 2024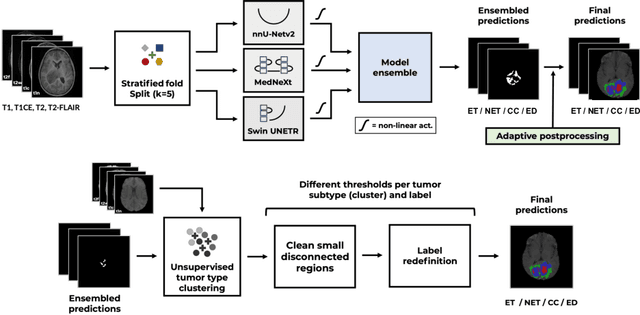
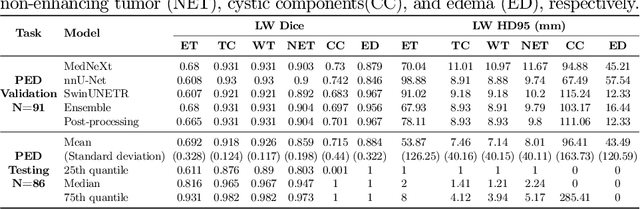
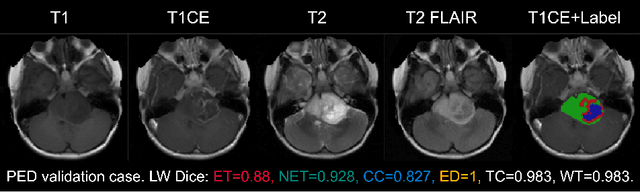
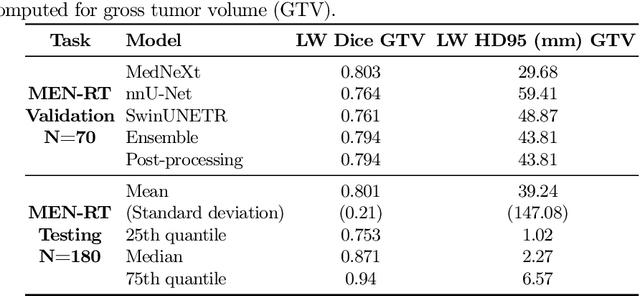
Abstract:Accurate and automatic segmentation of brain tumors in multi-parametric magnetic resonance imaging (mpMRI) is essential for quantitative measurements, which play an increasingly important role in clinical diagnosis and prognosis. The International Brain Tumor Segmentation (BraTS) Challenge 2024 offers a unique benchmarking opportunity, including various types of brain tumors in both adult and pediatric populations, such as pediatric brain tumors (PED), meningiomas (MEN-RT) and brain metastases (MET), among others. Compared to previous editions, BraTS 2024 has implemented changes to substantially increase clinical relevance, such as refined tumor regions for evaluation. We propose a deep learning-based ensemble approach that integrates state-of-the-art segmentation models. Additionally, we introduce innovative, adaptive pre- and post-processing techniques that employ MRI-based radiomic analyses to differentiate tumor subtypes. Given the heterogeneous nature of the tumors present in the BraTS datasets, this approach enhances the precision and generalizability of segmentation models. On the final testing sets, our method achieved mean lesion-wise Dice similarity coefficients of 0.926, 0.801, and 0.688 for the whole tumor in PED, MEN-RT, and MET, respectively. These results demonstrate the effectiveness of our approach in improving segmentation performance and generalizability for various brain tumor types.
Model Ensemble for Brain Tumor Segmentation in Magnetic Resonance Imaging
Sep 12, 2024



Abstract:Segmenting brain tumors in multi-parametric magnetic resonance imaging enables performing quantitative analysis in support of clinical trials and personalized patient care. This analysis provides the potential to impact clinical decision-making processes, including diagnosis and prognosis. In 2023, the well-established Brain Tumor Segmentation (BraTS) challenge presented a substantial expansion with eight tasks and 4,500 brain tumor cases. In this paper, we present a deep learning-based ensemble strategy that is evaluated for newly included tumor cases in three tasks: pediatric brain tumors (PED), intracranial meningioma (MEN), and brain metastases (MET). In particular, we ensemble outputs from state-of-the-art nnU-Net and Swin UNETR models on a region-wise basis. Furthermore, we implemented a targeted post-processing strategy based on a cross-validated threshold search to improve the segmentation results for tumor sub-regions. The evaluation of our proposed method on unseen test cases for the three tasks resulted in lesion-wise Dice scores for PED: 0.653, 0.809, 0.826; MEN: 0.876, 0.867, 0.849; and MET: 0.555, 0.6, 0.58; for the enhancing tumor, tumor core, and whole tumor, respectively. Our method was ranked first for PED, third for MEN, and fourth for MET, respectively.
Zero-Shot Pediatric Tuberculosis Detection in Chest X-Rays using Self-Supervised Learning
Feb 22, 2024



Abstract:Tuberculosis (TB) remains a significant global health challenge, with pediatric cases posing a major concern. The World Health Organization (WHO) advocates for chest X-rays (CXRs) for TB screening. However, visual interpretation by radiologists can be subjective, time-consuming and prone to error, especially in pediatric TB. Artificial intelligence (AI)-driven computer-aided detection (CAD) tools, especially those utilizing deep learning, show promise in enhancing lung disease detection. However, challenges include data scarcity and lack of generalizability. In this context, we propose a novel self-supervised paradigm leveraging Vision Transformers (ViT) for improved TB detection in CXR, enabling zero-shot pediatric TB detection. We demonstrate improvements in TB detection performance ($\sim$12.7% and $\sim$13.4% top AUC/AUPR gains in adults and children, respectively) when conducting self-supervised pre-training when compared to fully-supervised (i.e., non pre-trained) ViT models, achieving top performances of 0.959 AUC and 0.962 AUPR in adult TB detection, and 0.697 AUC and 0.607 AUPR in zero-shot pediatric TB detection. As a result, this work demonstrates that self-supervised learning on adult CXRs effectively extends to challenging downstream tasks such as pediatric TB detection, where data are scarce.
A Lightweight, Rapid and Efficient Deep Convolutional Network for Chest X-Ray Tuberculosis Detection
Sep 05, 2023



Abstract:Tuberculosis (TB) is still recognized as one of the leading causes of death worldwide. Recent advances in deep learning (DL) have shown to enhance radiologists' ability to interpret chest X-ray (CXR) images accurately and with fewer errors, leading to a better diagnosis of this disease. However, little work has been done to develop models capable of diagnosing TB that offer good performance while being efficient, fast and computationally inexpensive. In this work, we propose LightTBNet, a novel lightweight, fast and efficient deep convolutional network specially customized to detect TB from CXR images. Using a total of 800 frontal CXR images from two publicly available datasets, our solution yielded an accuracy, F1 and area under the ROC curve (AUC) of 0.906, 0.907 and 0.961, respectively, on an independent test subset. The proposed model demonstrates outstanding performance while delivering a rapid prediction, with minimal computational and memory requirements, making it highly suitable for deployment in handheld devices that can be used in low-resource areas with high TB prevalence. Code publicly available at https://github.com/dani-capellan/LightTBNet.
* 5 pages, 3 figures, 3 tables. This paper has been accepted at ISBI 2023
Deep learning-based lung segmentation and automatic regional template in chest X-ray images for pediatric tuberculosis
Jan 31, 2023Abstract:Tuberculosis (TB) is still considered a leading cause of death and a substantial threat to global child health. Both TB infection and disease are curable using antibiotics. However, most children who die of TB are never diagnosed or treated. In clinical practice, experienced physicians assess TB by examining chest X-rays (CXR). Pediatric CXR has specific challenges compared to adult CXR, which makes TB diagnosis in children more difficult. Computer-aided diagnosis systems supported by Artificial Intelligence have shown performance comparable to experienced radiologist TB readings, which could ease mass TB screening and reduce clinical burden. We propose a multi-view deep learning-based solution which, by following a proposed template, aims to automatically regionalize and extract lung and mediastinal regions of interest from pediatric CXR images where key TB findings may be present. Experimental results have shown accurate region extraction, which can be used for further analysis to confirm TB finding presence and severity assessment. Code publicly available at https://github.com/dani-capellan/pTB_LungRegionExtractor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge