Mario Ceresa
BCN MedTech, Dept. of Information and Communication Technologies, Universitat Pompeu Fabra, Barcelona, Spain
Retrieval Augmented Generation Evaluation for Health Documents
May 07, 2025Abstract:Safe and trustworthy use of Large Language Models (LLM) in the processing of healthcare documents and scientific papers could substantially help clinicians, scientists and policymakers in overcoming information overload and focusing on the most relevant information at a given moment. Retrieval Augmented Generation (RAG) is a promising method to leverage the potential of LLMs while enhancing the accuracy of their outcomes. This report assesses the potentials and shortcomings of such approaches in the automatic knowledge synthesis of different types of documents in the health domain. To this end, it describes: (1) an internally developed proof of concept pipeline that employs state-of-the-art practices to deliver safe and trustable analysis for healthcare documents and scientific papers called RAGEv (Retrieval Augmented Generation Evaluation); (2) a set of evaluation tools for LLM-based document retrieval and generation; (3) a benchmark dataset to verify the accuracy and veracity of the results called RAGEv-Bench. It concludes that careful implementations of RAG techniques could minimize most of the common problems in the use of LLMs for document processing in the health domain, obtaining very high scores both on short yes/no answers and long answers. There is a high potential for incorporating it into the day-to-day work of policy support tasks, but additional efforts are required to obtain a consistent and trustworthy tool.
Conformal Risk Control for Pulmonary Nodule Detection
Dec 28, 2024



Abstract:Quantitative tools are increasingly appealing for decision support in healthcare, driven by the growing capabilities of advanced AI systems. However, understanding the predictive uncertainties surrounding a tool's output is crucial for decision-makers to ensure reliable and transparent decisions. In this paper, we present a case study on pulmonary nodule detection for lung cancer screening, enhancing an advanced detection model with an uncertainty quantification technique called conformal risk control (CRC). We demonstrate that prediction sets with conformal guarantees are attractive measures of predictive uncertainty in the safety-critical healthcare domain, allowing end-users to achieve arbitrary validity by trading off false positives and providing formal statistical guarantees on model performance. Among ground-truth nodules annotated by at least three radiologists, our model achieves a sensitivity that is competitive with that generally achieved by individual radiologists, with a slight increase in false positives. Furthermore, we illustrate the risks of using off-the-shelve prediction models when faced with ontological uncertainty, such as when radiologists disagree on what constitutes the ground truth on pulmonary nodules.
Epidemic Information Extraction for Event-Based Surveillance using Large Language Models
Aug 26, 2024Abstract:This paper presents a novel approach to epidemic surveillance, leveraging the power of Artificial Intelligence and Large Language Models (LLMs) for effective interpretation of unstructured big data sources, like the popular ProMED and WHO Disease Outbreak News. We explore several LLMs, evaluating their capabilities in extracting valuable epidemic information. We further enhance the capabilities of the LLMs using in-context learning, and test the performance of an ensemble model incorporating multiple open-source LLMs. The findings indicate that LLMs can significantly enhance the accuracy and timeliness of epidemic modelling and forecasting, offering a promising tool for managing future pandemic events.
* 11 pages, 4 figures, Ninth International Congress on Information and Communication Technology (ICICT 2024)
Unsupervised Segmentation of Fetal Brain MRI using Deep Learning Cascaded Registration
Jul 07, 2023



Abstract:Accurate segmentation of fetal brain magnetic resonance images is crucial for analyzing fetal brain development and detecting potential neurodevelopmental abnormalities. Traditional deep learning-based automatic segmentation, although effective, requires extensive training data with ground-truth labels, typically produced by clinicians through a time-consuming annotation process. To overcome this challenge, we propose a novel unsupervised segmentation method based on multi-atlas segmentation, that accurately segments multiple tissues without relying on labeled data for training. Our method employs a cascaded deep learning network for 3D image registration, which computes small, incremental deformations to the moving image to align it precisely with the fixed image. This cascaded network can then be used to register multiple annotated images with the image to be segmented, and combine the propagated labels to form a refined segmentation. Our experiments demonstrate that the proposed cascaded architecture outperforms the state-of-the-art registration methods that were tested. Furthermore, the derived segmentation method achieves similar performance and inference time to nnU-Net while only using a small subset of annotated data for the multi-atlas segmentation task and none for training the network. Our pipeline for registration and multi-atlas segmentation is publicly available at https://github.com/ValBcn/CasReg.
An Uncertainty-aware Hierarchical Probabilistic Network for Early Prediction, Quantification and Segmentation of Pulmonary Tumour Growth
Apr 18, 2021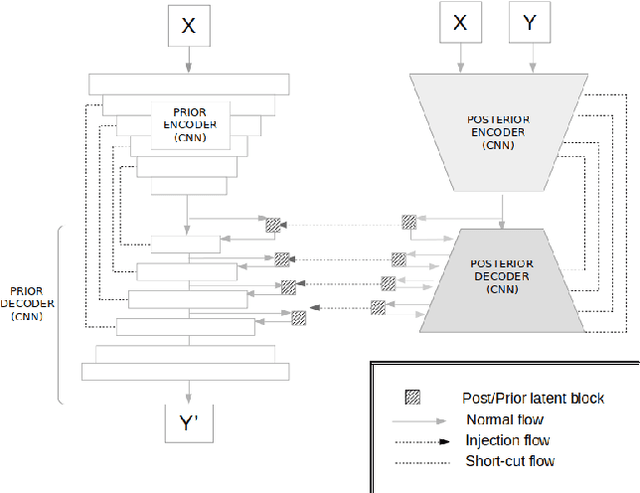
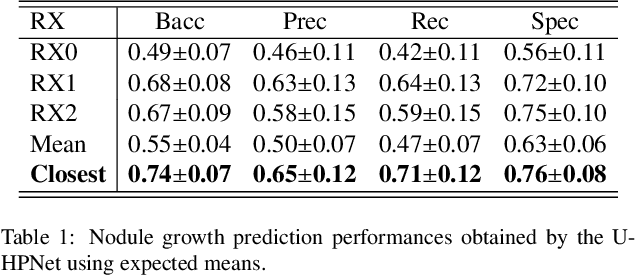
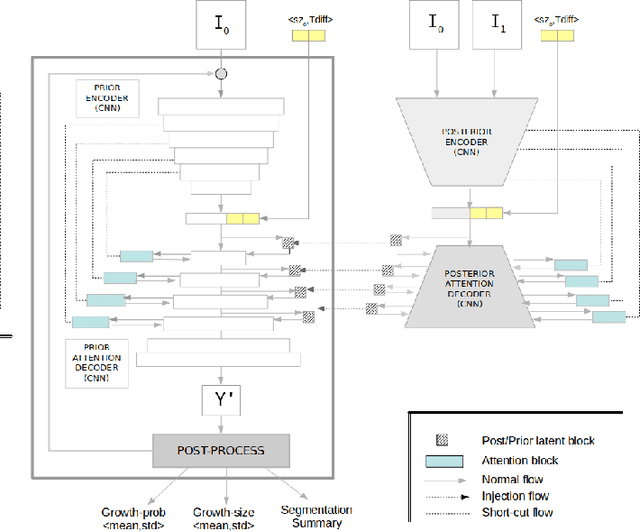
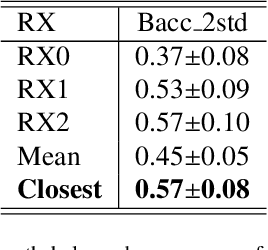
Abstract:Early detection and quantification of tumour growth would help clinicians to prescribe more accurate treatments and provide better surgical planning. However, the multifactorial and heterogeneous nature of lung tumour progression hampers identification of growth patterns. In this study, we present a novel method based on a deep hierarchical generative and probabilistic framework that, according to radiological guidelines, predicts tumour growth, quantifies its size and provides a semantic appearance of the future nodule. Unlike previous deterministic solutions, the generative characteristic of our approach also allows us to estimate the uncertainty in the predictions, especially important for complex and doubtful cases. Results of evaluating this method on an independent test set reported a tumour growth balanced accuracy of 74%, a tumour growth size MAE of 1.77 mm and a tumour segmentation Dice score of 78%. These surpassed the performances of equivalent deterministic and alternative generative solutions (i.e. probabilistic U-Net, Bayesian test dropout and Pix2Pix GAN) confirming the suitability of our approach.
Detection, growth quantification and malignancy prediction of pulmonary nodules using deep convolutional networks in follow-up CT scans
Mar 26, 2021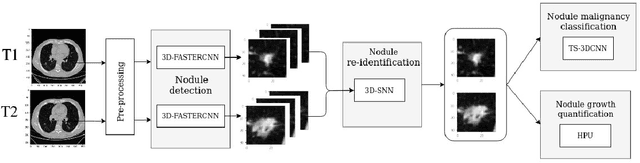
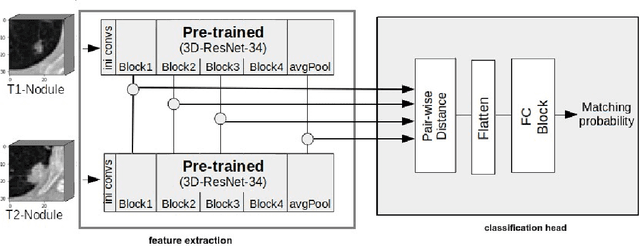
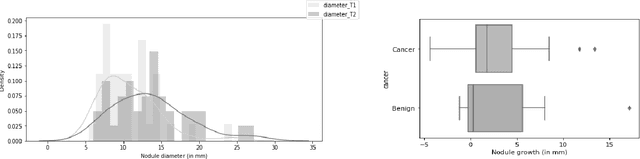
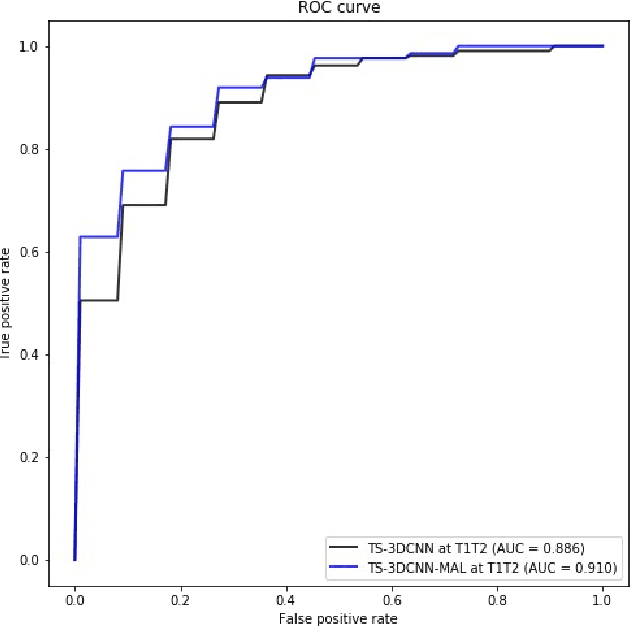
Abstract:We address the problem of supporting radiologists in the longitudinal management of lung cancer. Therefore, we proposed a deep learning pipeline, composed of four stages that completely automatized from the detection of nodules to the classification of cancer, through the detection of growth in the nodules. In addition, the pipeline integrated a novel approach for nodule growth detection, which relied on a recent hierarchical probabilistic U-Net adapted to report uncertainty estimates. Also, a second novel method was introduced for lung cancer nodule classification, integrating into a two stream 3D-CNN network the estimated nodule malignancy probabilities derived from a pretrained nodule malignancy network. The pipeline was evaluated in a longitudinal cohort and reported comparable performances to the state of art.
Pulmonary Nodule Malignancy Classification Using its Temporal Evolution with Two-Stream 3D Convolutional Neural Networks
May 22, 2020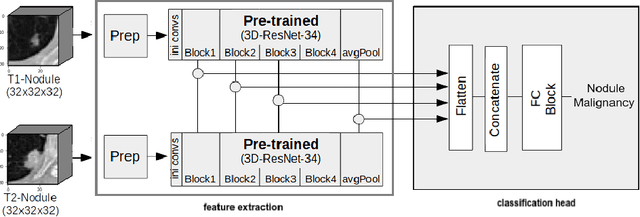
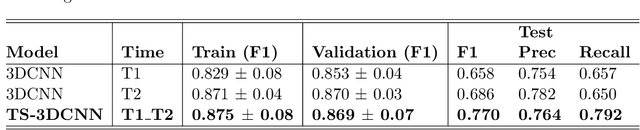
Abstract:Nodule malignancy assessment is a complex, time-consuming and error-prone task. Current clinical practice requires measuring changes in size and density of the nodule at different time-points. State of the art solutions rely on 3D convolutional neural networks built on pulmonary nodules obtained from single CT scan per patient. In this work, we propose a two-stream 3D convolutional neural network that predicts malignancy by jointly analyzing two pulmonary nodule volumes from the same patient taken at different time-points. Best results achieve 77% of F1-score in test with an increment of 9% and 12% of F1-score with respect to the same network trained with images from a single time-point.
Re-Identification and Growth Detection of Pulmonary Nodules without Image Registration Using 3D Siamese Neural Networks
Dec 22, 2019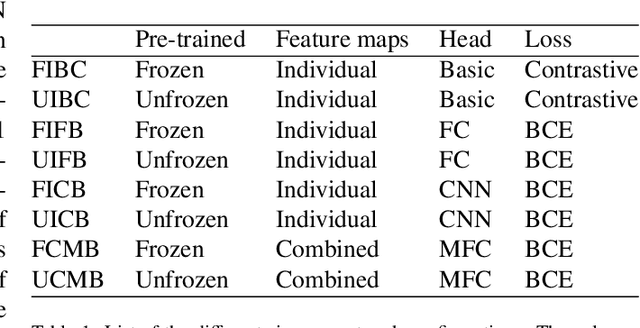
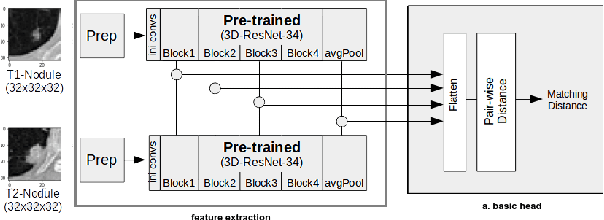
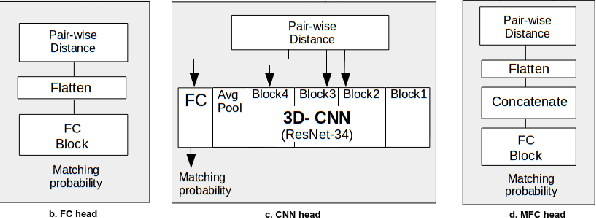
Abstract:Lung cancer follow-up is a complex, error prone, and time consuming task for clinical radiologists. Several lung CT scan images taken at different time points of a given patient need to be individually inspected, looking for possible cancerogenous nodules. Radiologists mainly focus their attention in nodule size, density, and growth to assess the existence of malignancy. In this study, we present a novel method based on a 3D siamese neural network, for the re-identification of nodules in a pair of CT scans of the same patient without the need for image registration. The network was integrated into a two-stage automatic pipeline to detect, match, and predict nodule growth given pairs of CT scans. Results on an independent test set reported a nodule detection sensitivity of 94.7%, an accuracy for temporal nodule matching of 88.8%, and a sensitivity of 92.0% with a precision of 88.4% for nodule growth detection.
Integration of Convolutional Neural Networks for Pulmonary Nodule Malignancy Assessment in a Lung Cancer Classification Pipeline
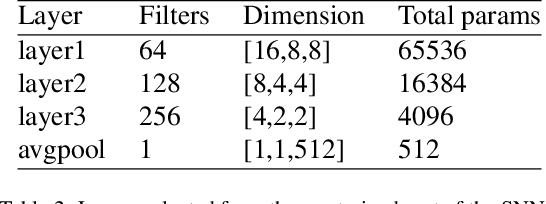
Dec 18, 2019
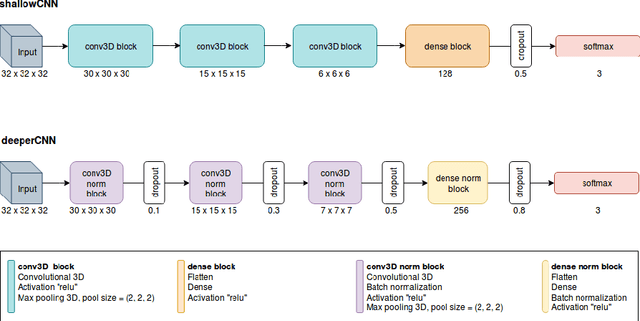
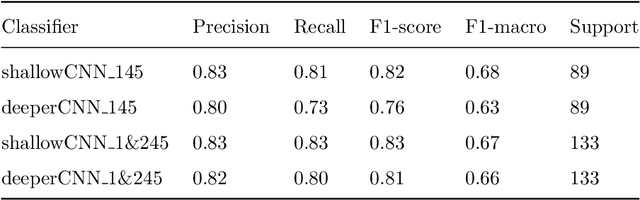
Abstract:The early identification of malignant pulmonary nodules is critical for better lung cancer prognosis and less invasive chemo or radio therapies. Nodule malignancy assessment done by radiologists is extremely useful for planning a preventive intervention but is, unfortunately, a complex, time-consuming and error-prone task. This explains the lack of large datasets containing radiologists malignancy characterization of nodules. In this article, we propose to assess nodule malignancy through 3D convolutional neural networks and to integrate it in an automated end-to-end existing pipeline of lung cancer detection. For training and testing purposes we used independent subsets of the LIDC dataset. Adding the probabilities of nodules malignity in a baseline lung cancer pipeline improved its F1-weighted score by 14.7%, whereas integrating the malignancy model itself using transfer learning outperformed the baseline prediction by 11.8% of F1-weighted score. Despite the limited size of the lung cancer datasets, integrating predictive models of nodule malignancy improves prediction of lung cancer.
* 26 pages, 5 figures
Fully automatic detection and segmentation of abdominal aortic thrombus in post-operative CTA images using deep convolutional neural networks
Apr 01, 2018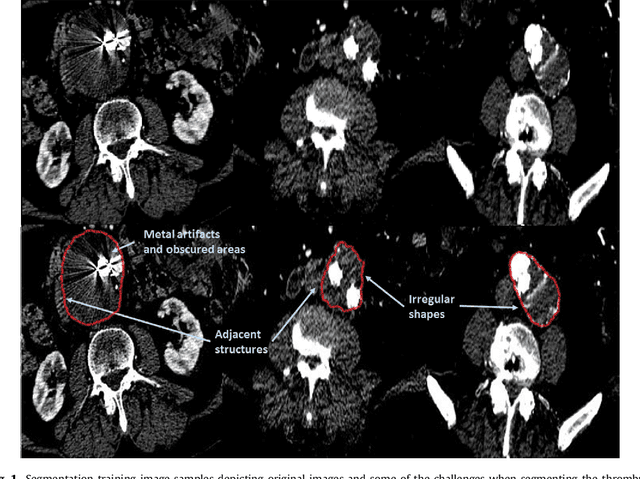
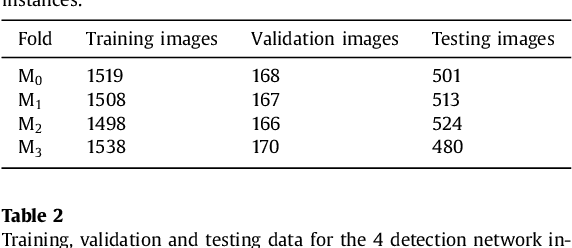
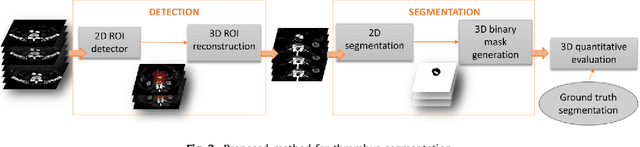

Abstract:Computerized Tomography Angiography (CTA) based follow-up of Abdominal Aortic Aneurysms (AAA) treated with Endovascular Aneurysm Repair (EVAR) is essential to evaluate the progress of the patient and detect complications. In this context, accurate quantification of post-operative thrombus volume is required. However, a proper evaluation is hindered by the lack of automatic, robust and reproducible thrombus segmentation algorithms. We propose a new fully automatic approach based on Deep Convolutional Neural Networks (DCNN) for robust and reproducible thrombus region of interest detection and subsequent fine thrombus segmentation. The DetecNet detection network is adapted to perform region of interest extraction from a complete CTA and a new segmentation network architecture, based on Fully Convolutional Networks and a Holistically-Nested Edge Detection Network, is presented. These networks are trained, validated and tested in 13 post-operative CTA volumes of different patients using a 4-fold cross-validation approach to provide more robustness to the results. Our pipeline achieves a Dice score of more than 82% for post-operative thrombus segmentation and provides a mean relative volume difference between ground truth and automatic segmentation that lays within the experienced human observer variance without the need of human intervention in most common cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge